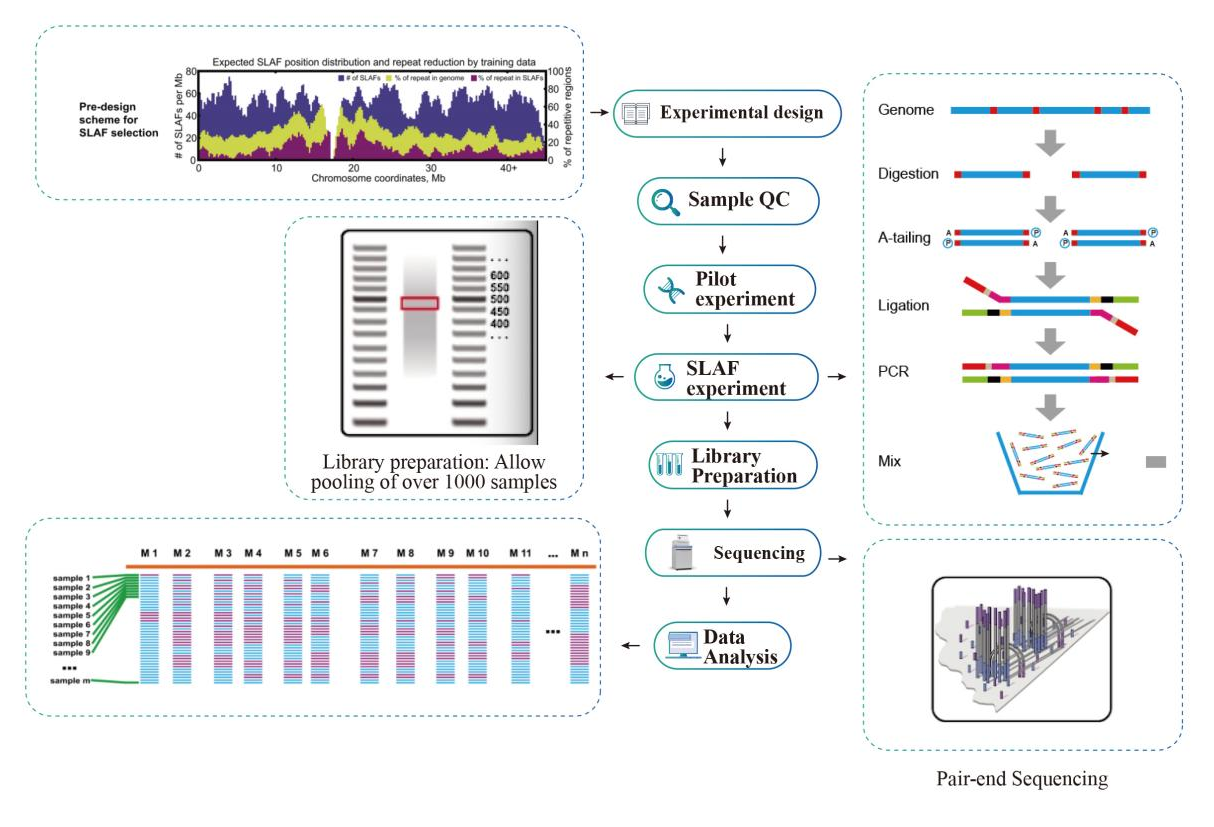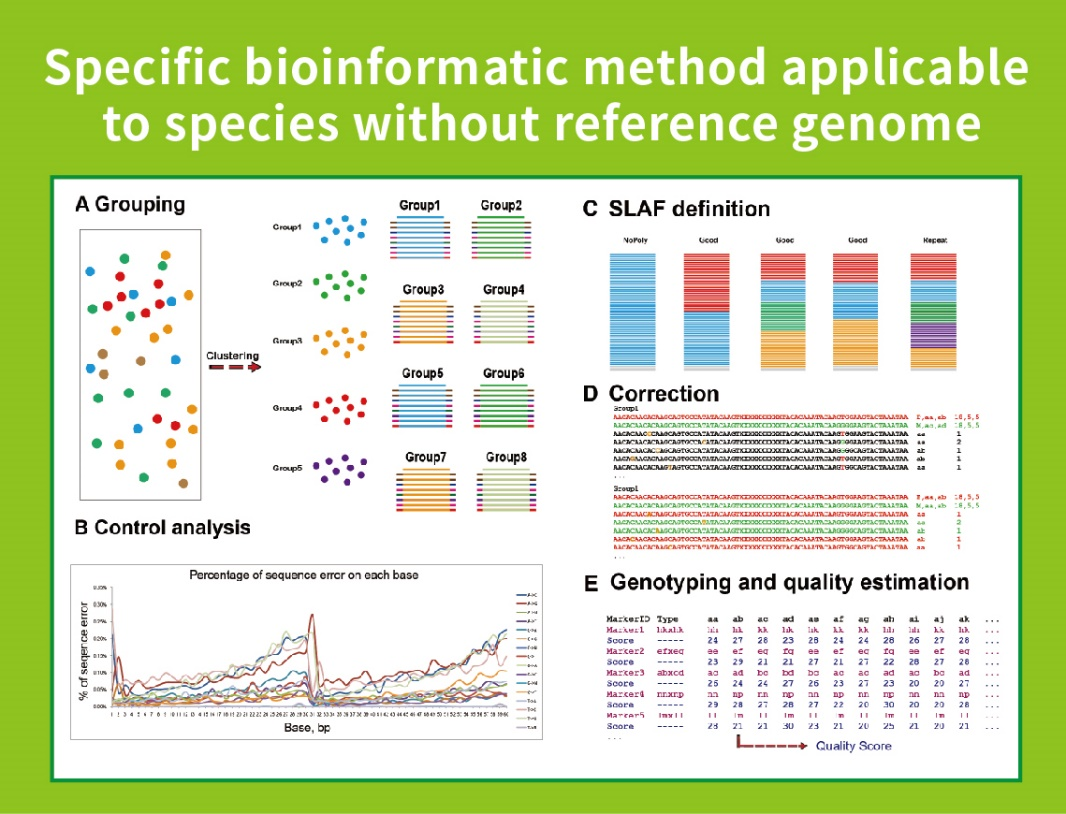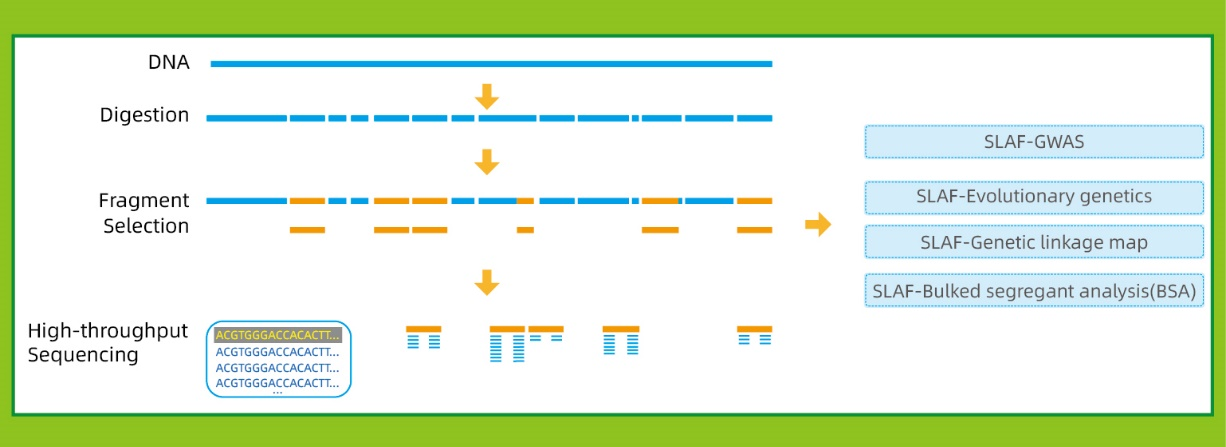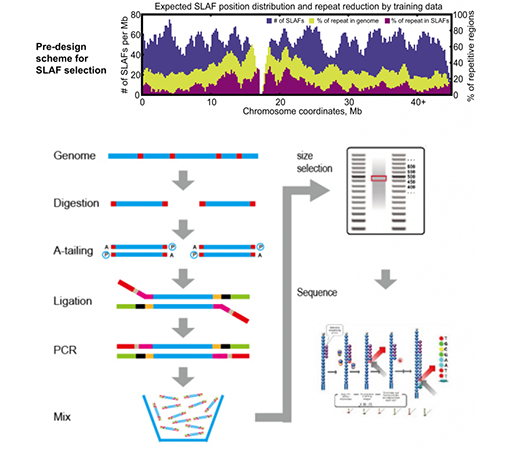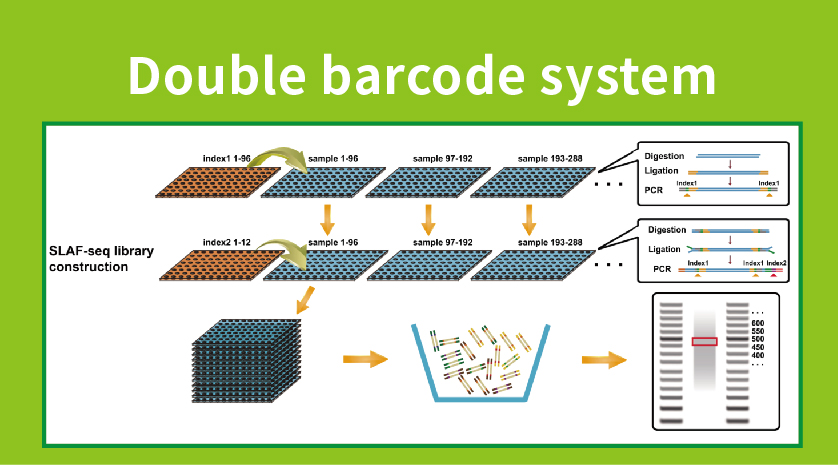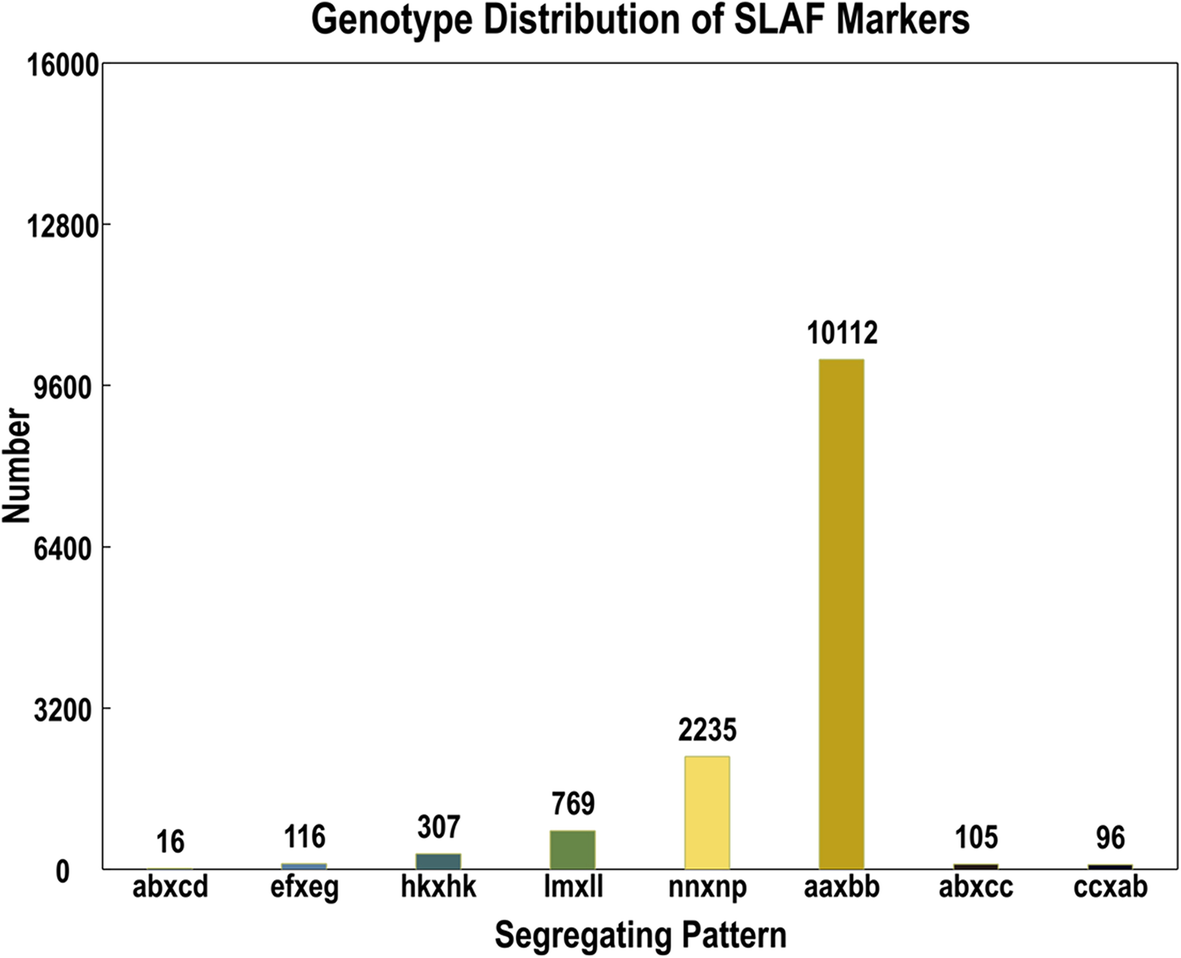
A high-density genetic map developed by specific-locus amplified fragment ( SLAF) sequencing and identification of a locus controlling anthocyanin pigmentation in stalk of Zicaitai (Brassica rapa L. ssp. chinensis var. purpurea) | BMC
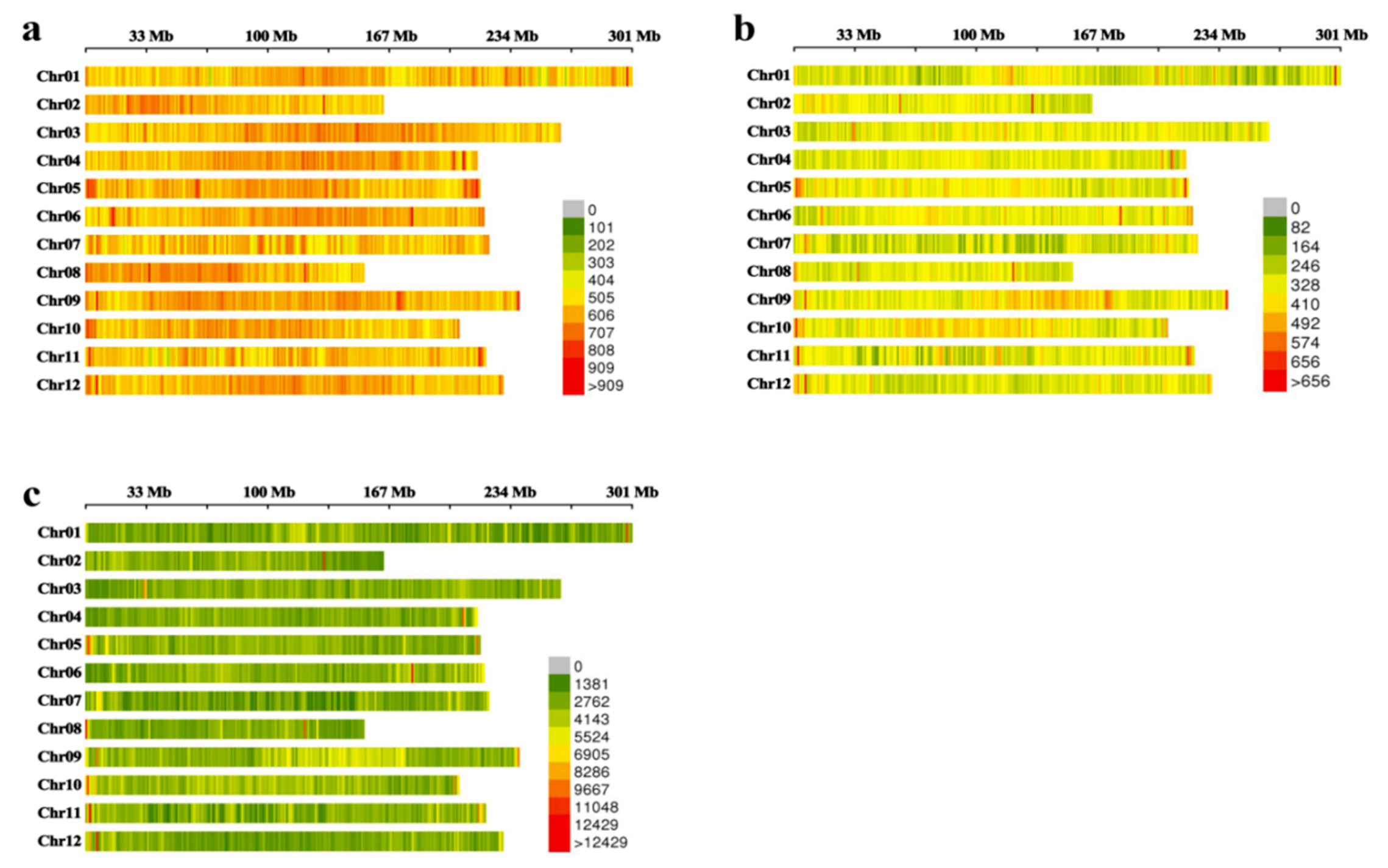
IJMS | Free Full-Text | Genome-Wide Correlation of 36 Agronomic Traits in the 287 Pepper (Capsicum) Accessions Obtained from the SLAF-seq-Based GWAS

A simplified scheme for SLAF-Seq. (a) Thirty lines with largest TGW and... | Download Scientific Diagram
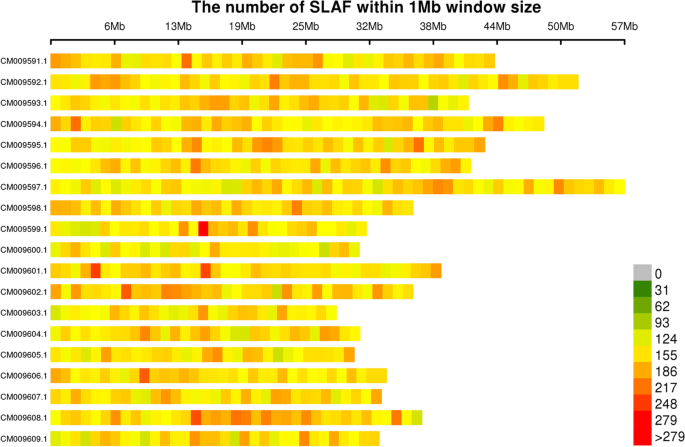
Investigation of genetic relationships within three Miscanthus species using SNP markers identified with SLAF-seq | BMC Genomics | Full Text
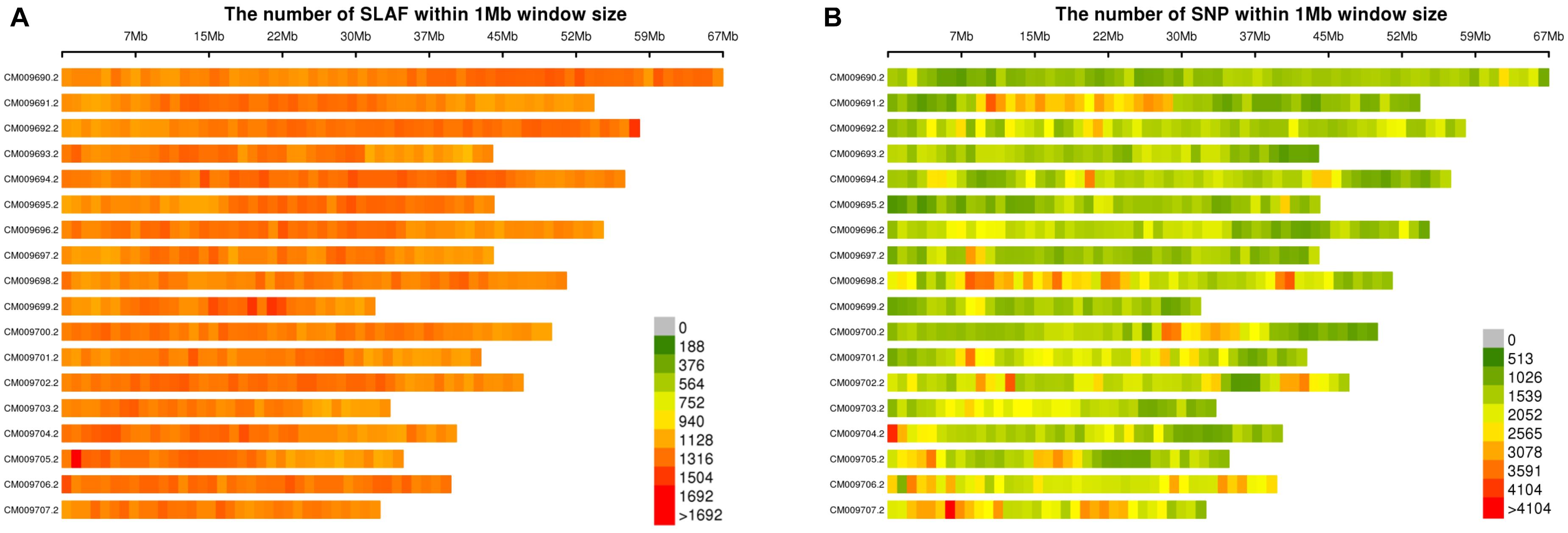
Frontiers | Genetic Divergence and Population Structure in Weedy and Cultivated Broomcorn Millets (Panicum miliaceum L.) Revealed by Specific-Locus Amplified Fragment Sequencing (SLAF-Seq)

SLAF-based high-density genetic map construction and QTL mapping for major economic traits in sea urchin Strongylocentrotus intermedius | Scientific Reports

Agronomy | Free Full-Text | An SNP-Based High-Density Genetic Linkage Map for Tetraploid Potato Using Specific Length Amplified Fragment Sequencing ( SLAF-Seq) Technology
SLAF-seq: An Efficient Method of Large-Scale De Novo SNP Discovery and Genotyping Using High-Throughput Sequencing | PLOS ONE

Specific-Locus Amplified Fragment Sequencing (SLAF-Seq) as High-Throughput SNP Genotyping Methods | SpringerLink
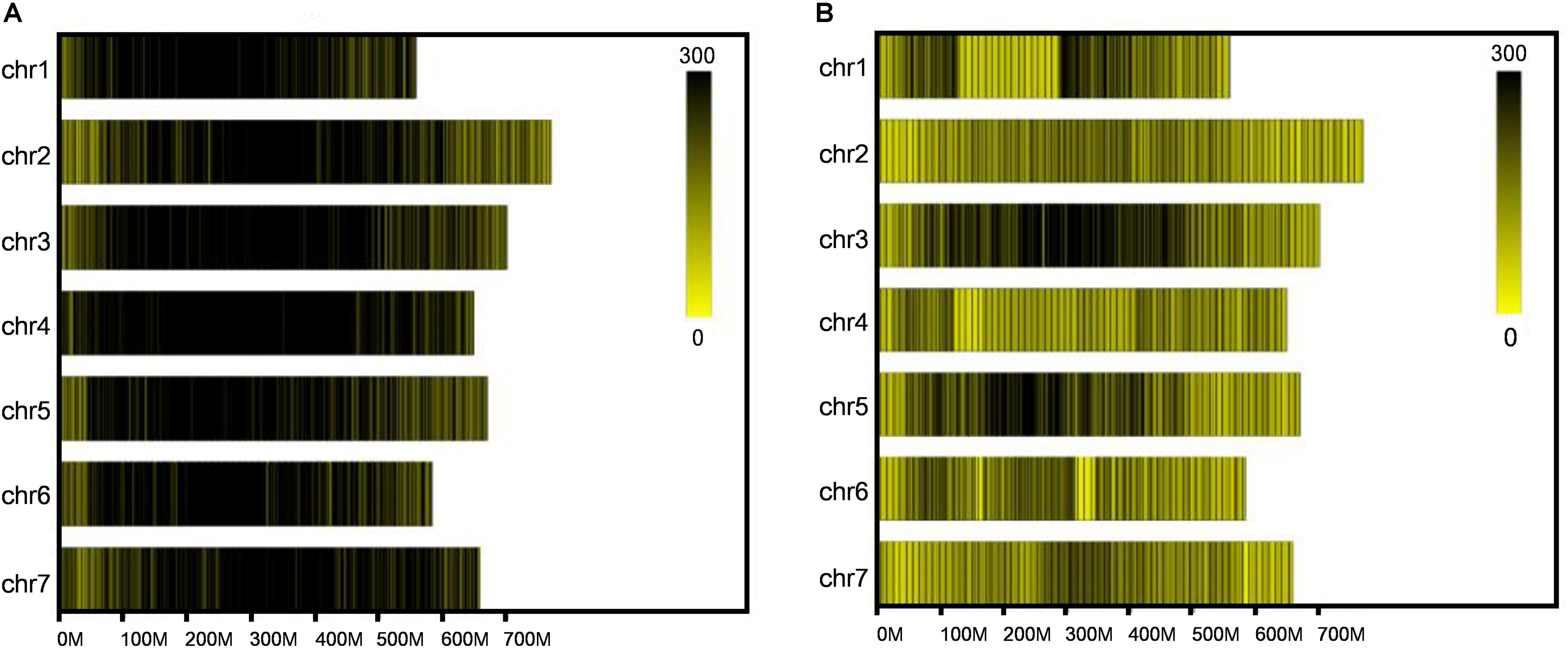
Frontiers | A High-Density Genetic Linkage Map of SLAFs and QTL Analysis of Grain Size and Weight in Barley (Hordeum vulgare L.)

An evaluation of genotyping by sequencing (GBS) to map the Breviaristatum-e (ari-e) locus in cultivated barley – topic of research paper in Biological sciences. Download scholarly article PDF and read for free

Genes | Free Full-Text | Molecular-Assisted Distinctness and Uniformity Testing Using SLAF-Sequencing Approach in Soybean

Construction of a High-Density Genetic Map Based on Large-Scale Marker Development in Coix lacryma-jobi L. Using Specifific-Locus Amplifified Fragment Sequencing (SLAF-seq) | bioRxiv
SLAF-seq: An Efficient Method of Large-Scale De Novo SNP Discovery and Genotyping Using High-Throughput Sequencing | PLOS ONE
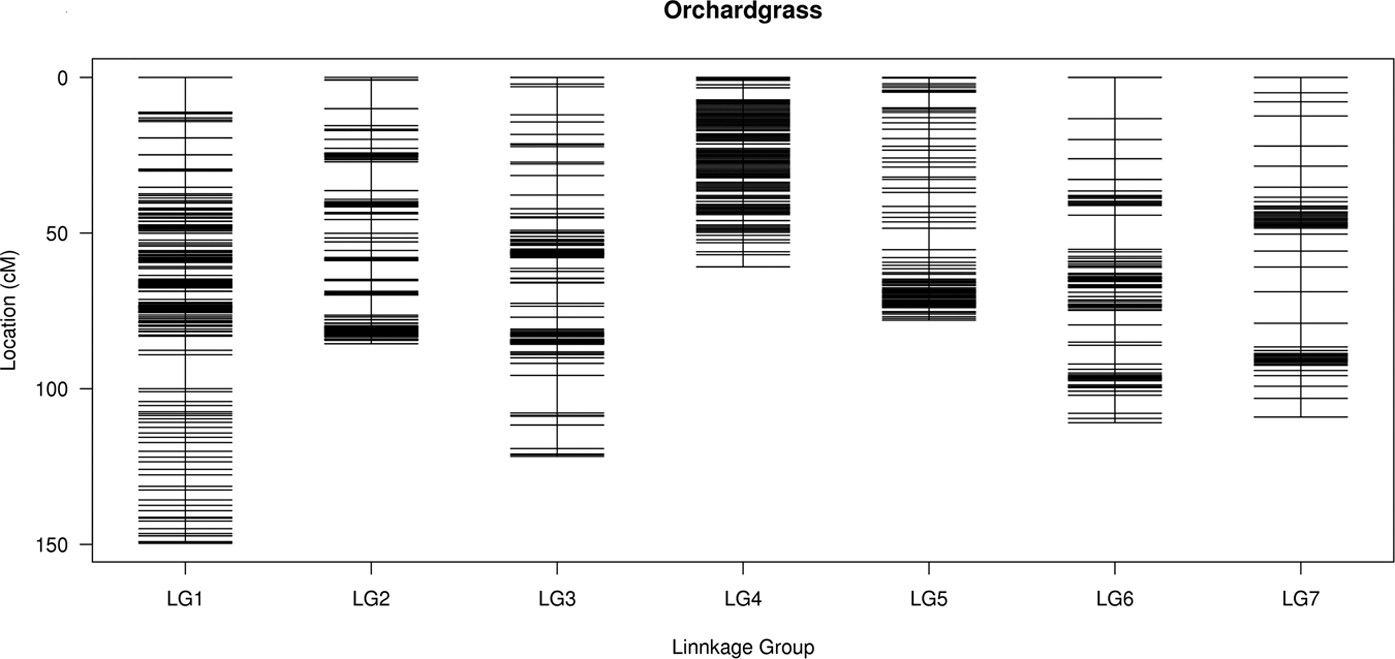
Construction of high-density genetic linkage map and identification of flowering-time QTLs in orchardgrass using SSRs and SLAF-seq | Scientific Reports

Specific-Locus Amplified Fragment Sequencing (SLAF-Seq) as High-Throughput SNP Genotyping Methods | SpringerLink

Genome-wide identification of meiotic recombination hot spots detected by SLAF in peanut (Arachis hypogaea L.) | Scientific Reports