INSPIIRED: A Pipeline for Quantitative Analysis of Sites of New DNA Integration in Cellular Genomes: Molecular Therapy - Methods & Clinical Development
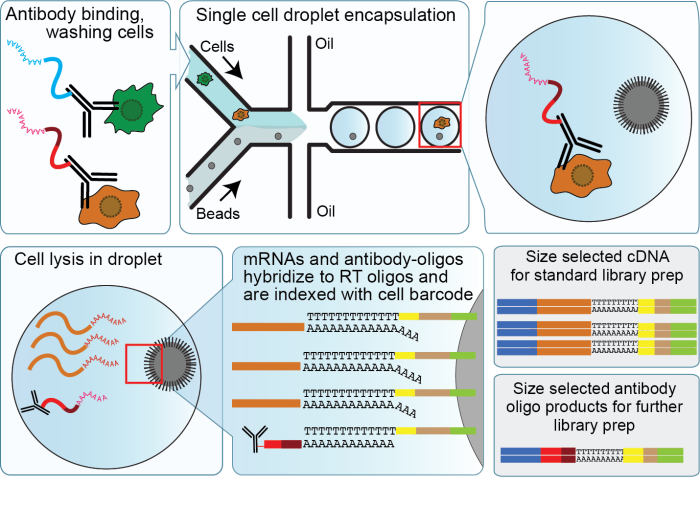
Precise genomic mapping of 5-hydroxymethylcytosine via covalent tether-directed sequencing | PLOS Biology

Schematic representation of integration site sequencing. Sonication... | Download Scientific Diagram

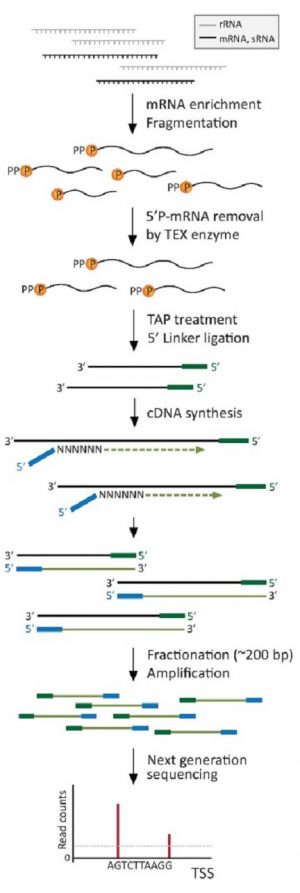
End-seq – map both 5' and 3' transcript ends genome-wide at single nucleotide resolution in 3 days | RNA-Seq Blog
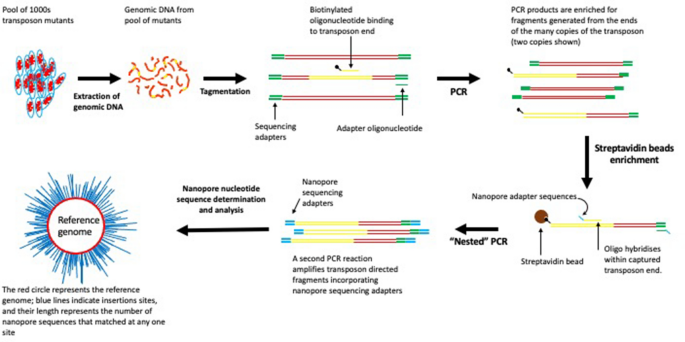
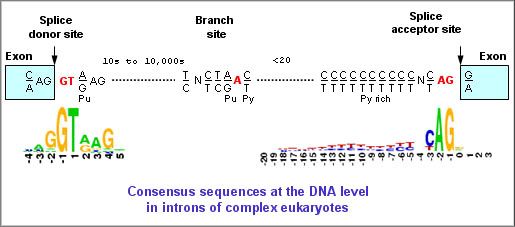
Long-read sequencing for identification of insertion sites in large transposon mutant libraries | Scientific Reports

Mapping of transposon directed insertion site sequences to repeated... | Download Scientific Diagram
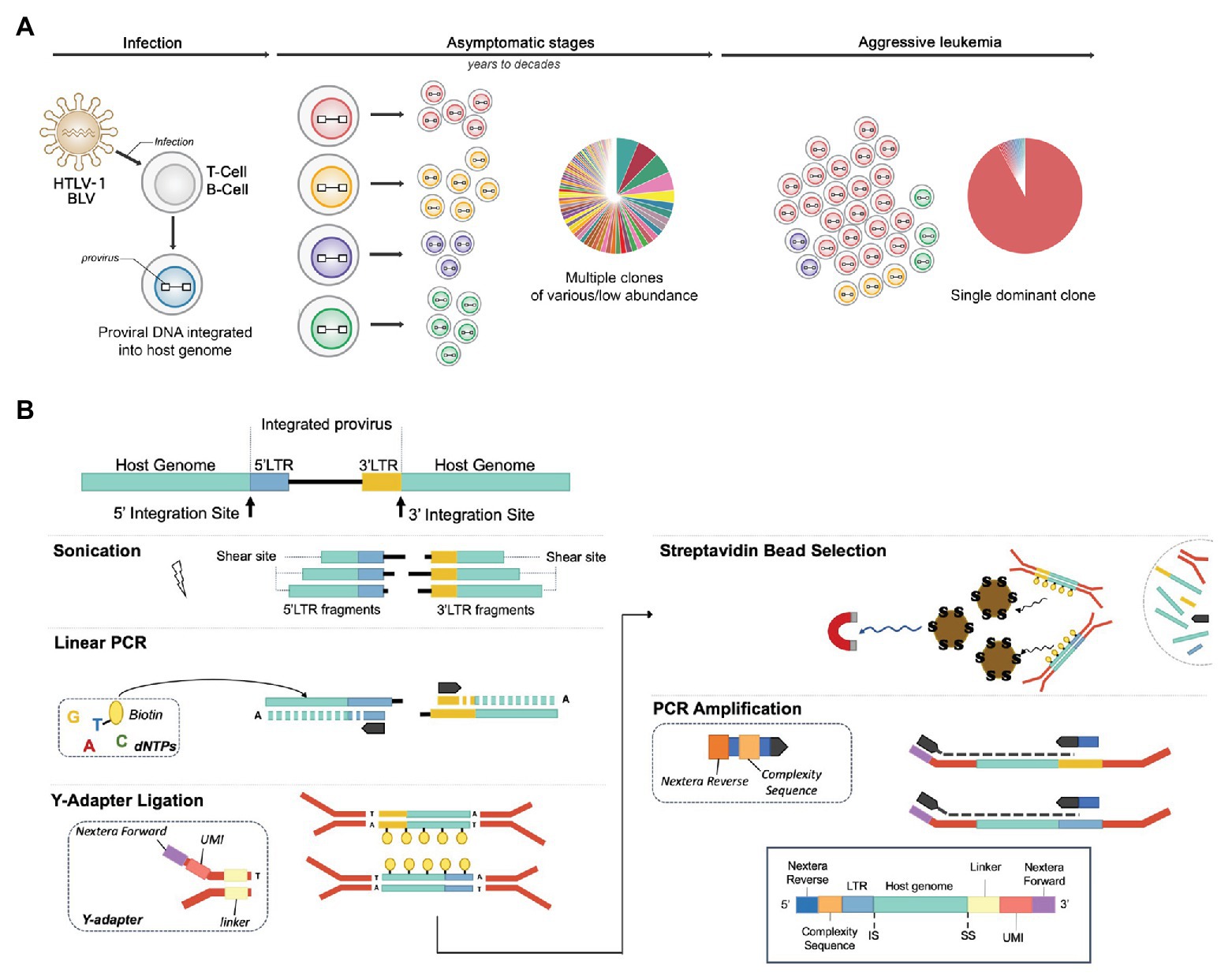
Frontiers | An Improved Sequencing-Based Bioinformatics Pipeline to Track the Distribution and Clonal Architecture of Proviral Integration Sites
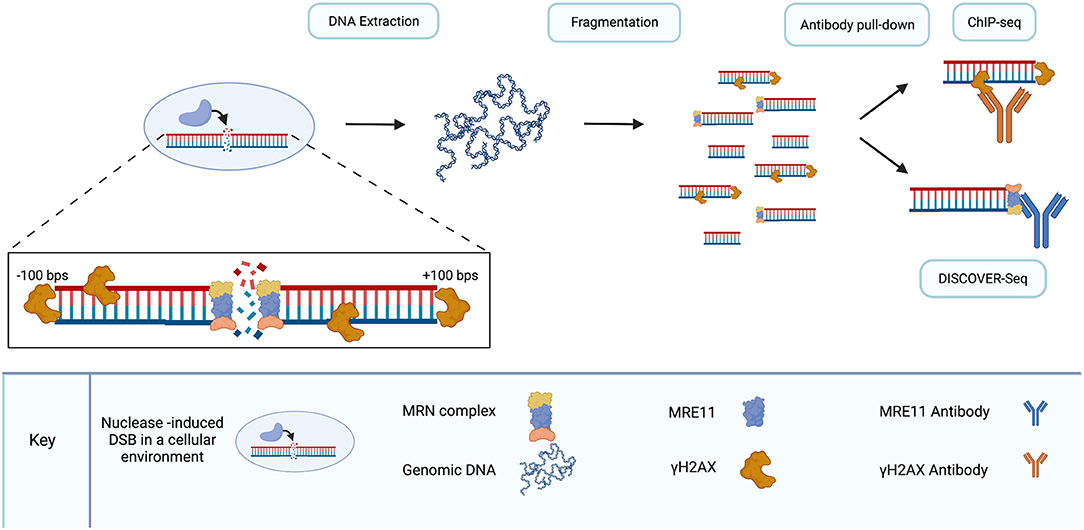
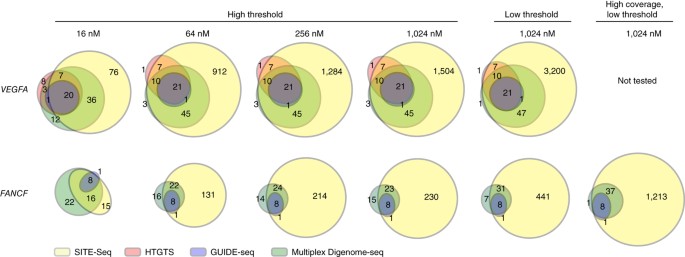
Frontiers | Off-Target Analysis in Gene Editing and Applications for Clinical Translation of CRISPR/Cas9 in HIV-1 Therapy

Initiation site sequencing (ini-seq). Schematic representation of the... | Download Scientific Diagram

CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR–Cas9 nuclease off-targets | Nature Methods

The CIRCLE-seq target enrichment strategy creates an in vitro library... | Download Scientific Diagram
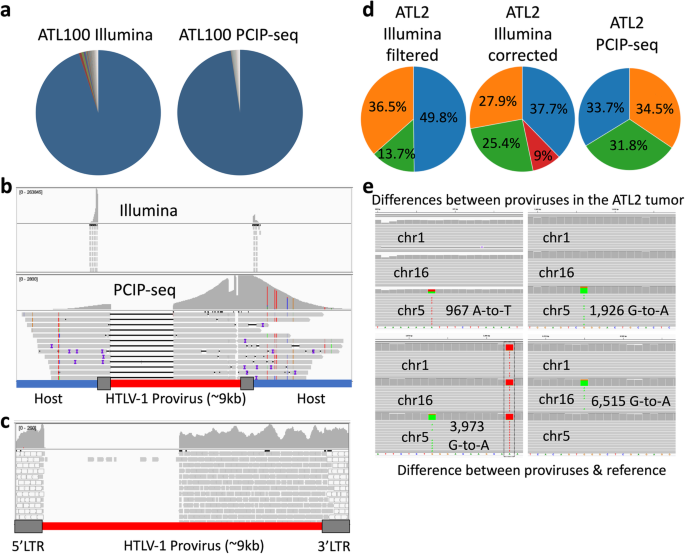
PCIP-seq: simultaneous sequencing of integrated viral genomes and their insertion sites with long reads | Genome Biology | Full Text

RADAR-seq: A RAre DAmage and Repair sequencing method for detecting DNA damage on a genome-wide scale - ScienceDirect

Combined HIV-1 sequence and integration site analysis informs viral dynamics and allows reconstruction of replicating viral ancestors | PNAS

HIV Integration Site Analysis of Cellular Models of HIV Latency with a Probe-Enriched Next-Generation Sequencing Assay | Journal of Virology
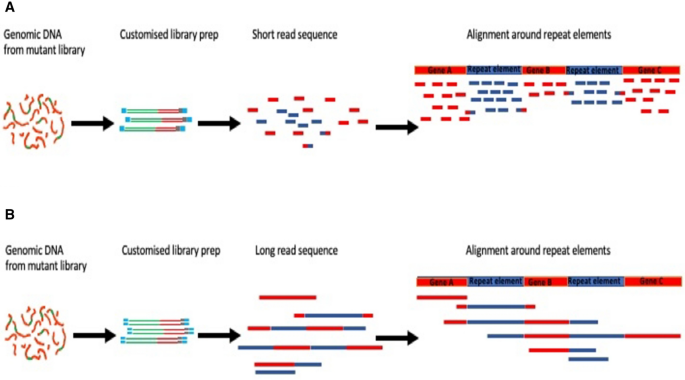
Long-read sequencing for identification of insertion sites in large transposon mutant libraries | Scientific Reports

CReVIS-Seq: A highly accurate and multiplexable method for genome-wide mapping of lentiviral integration sites: Molecular Therapy - Methods & Clinical Development