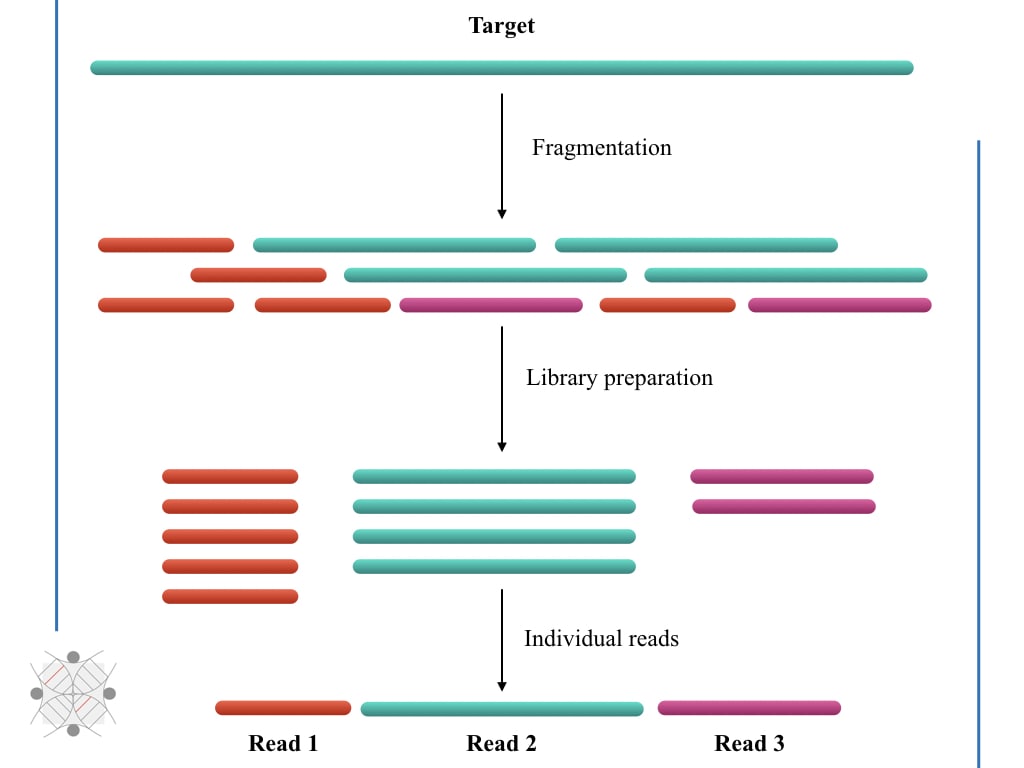
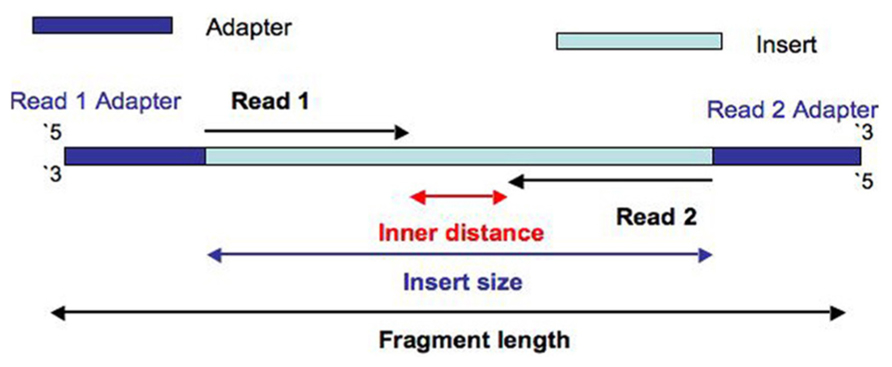
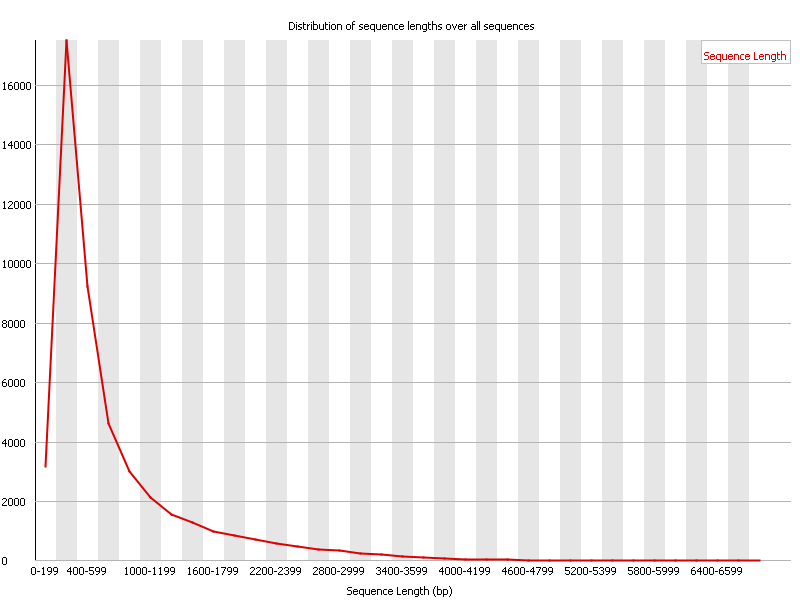
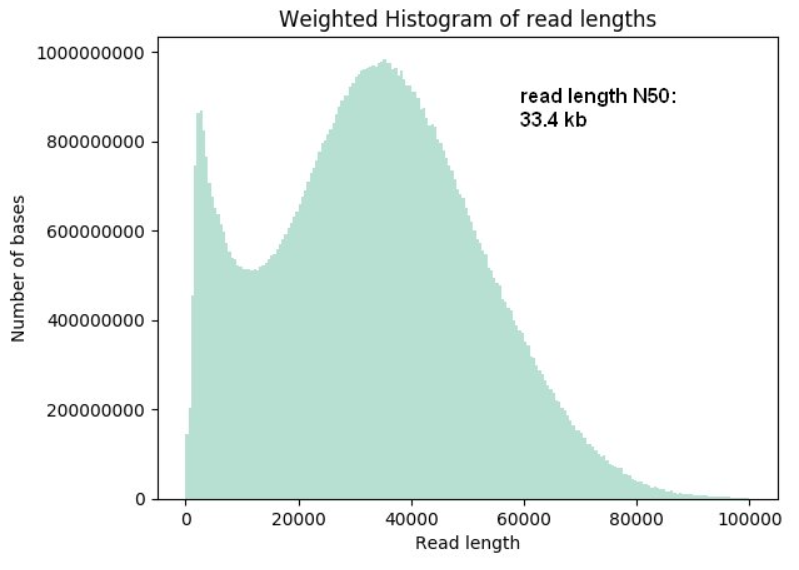
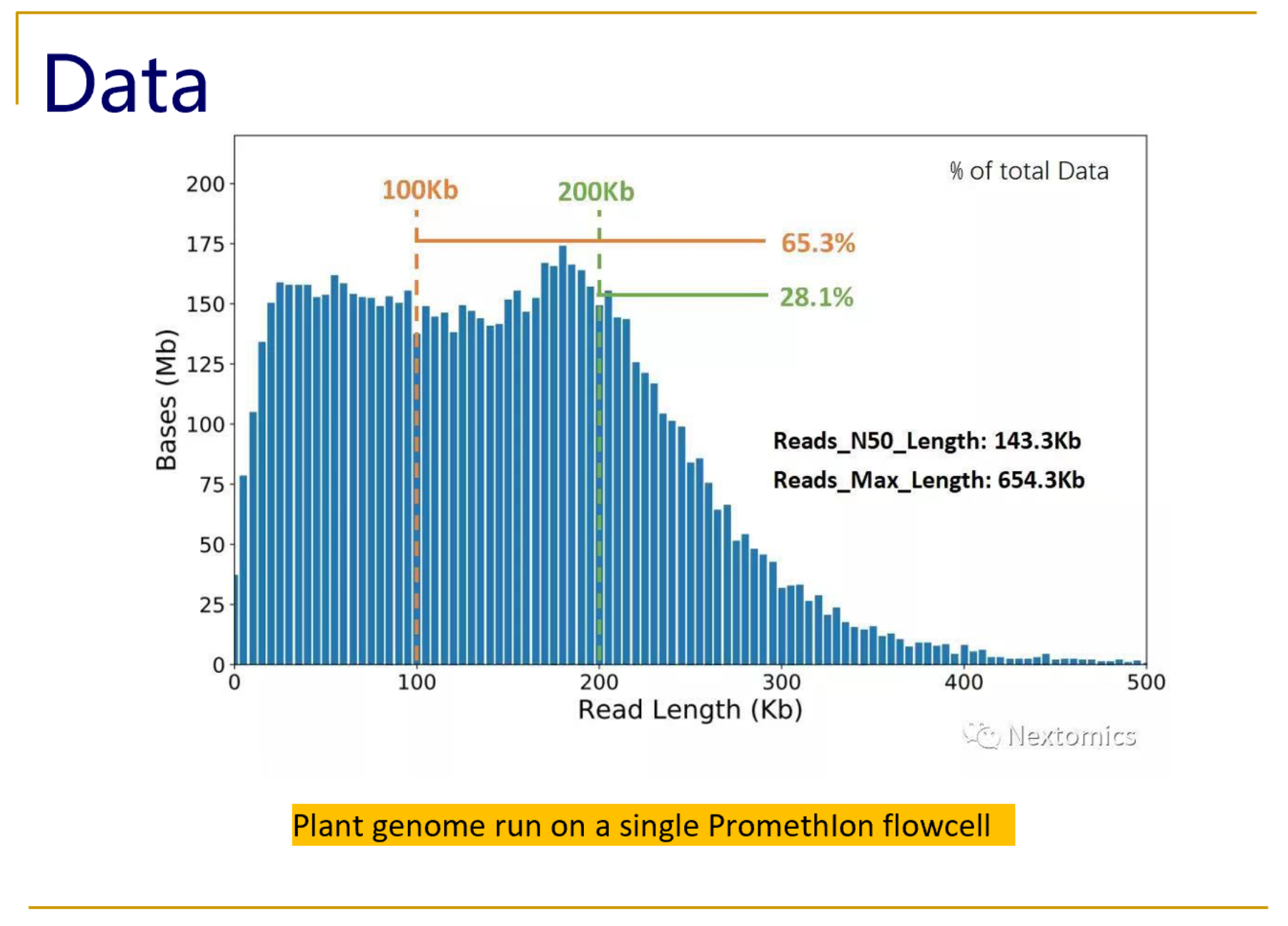
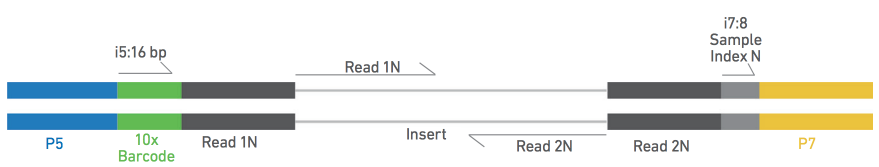
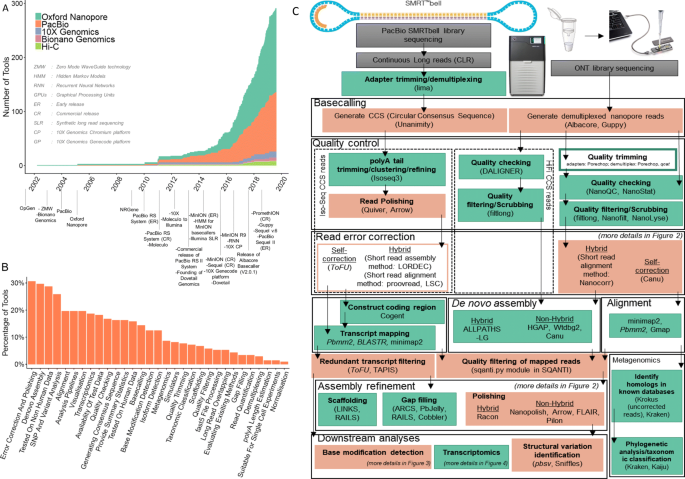
Unusual difference between "Demultiplexed sequence length summary" and " Sequencing company: read length" - User Support - QIIME 2 Forum

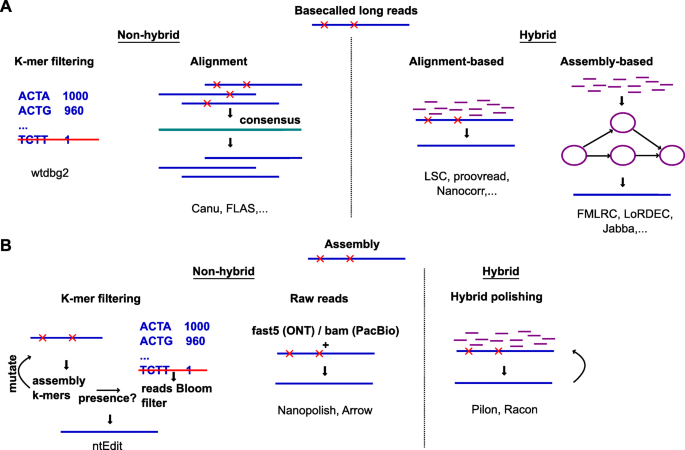
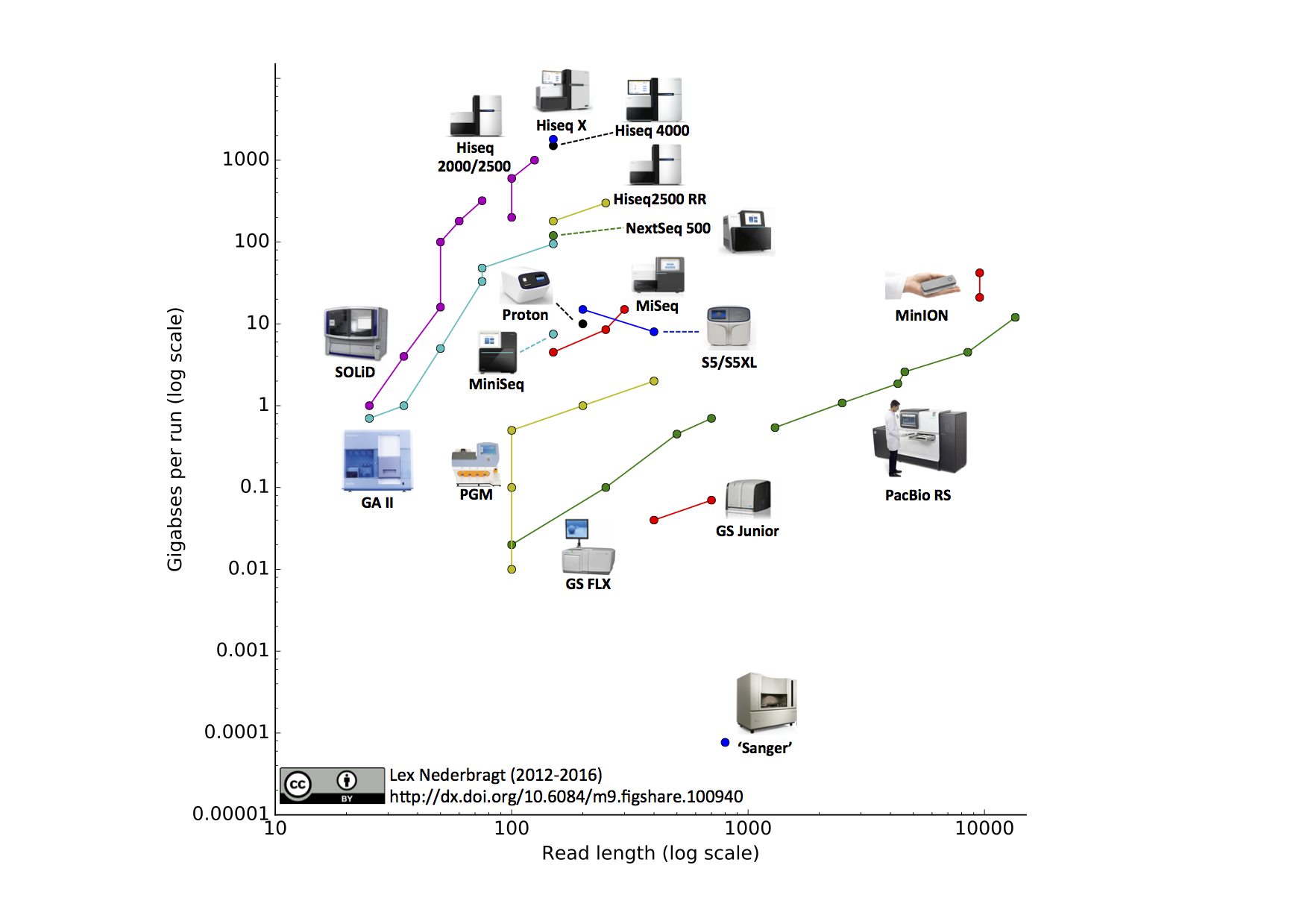
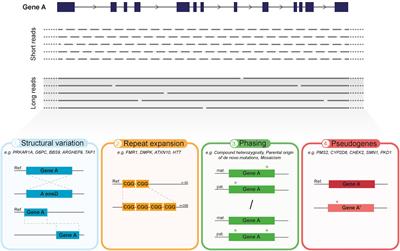
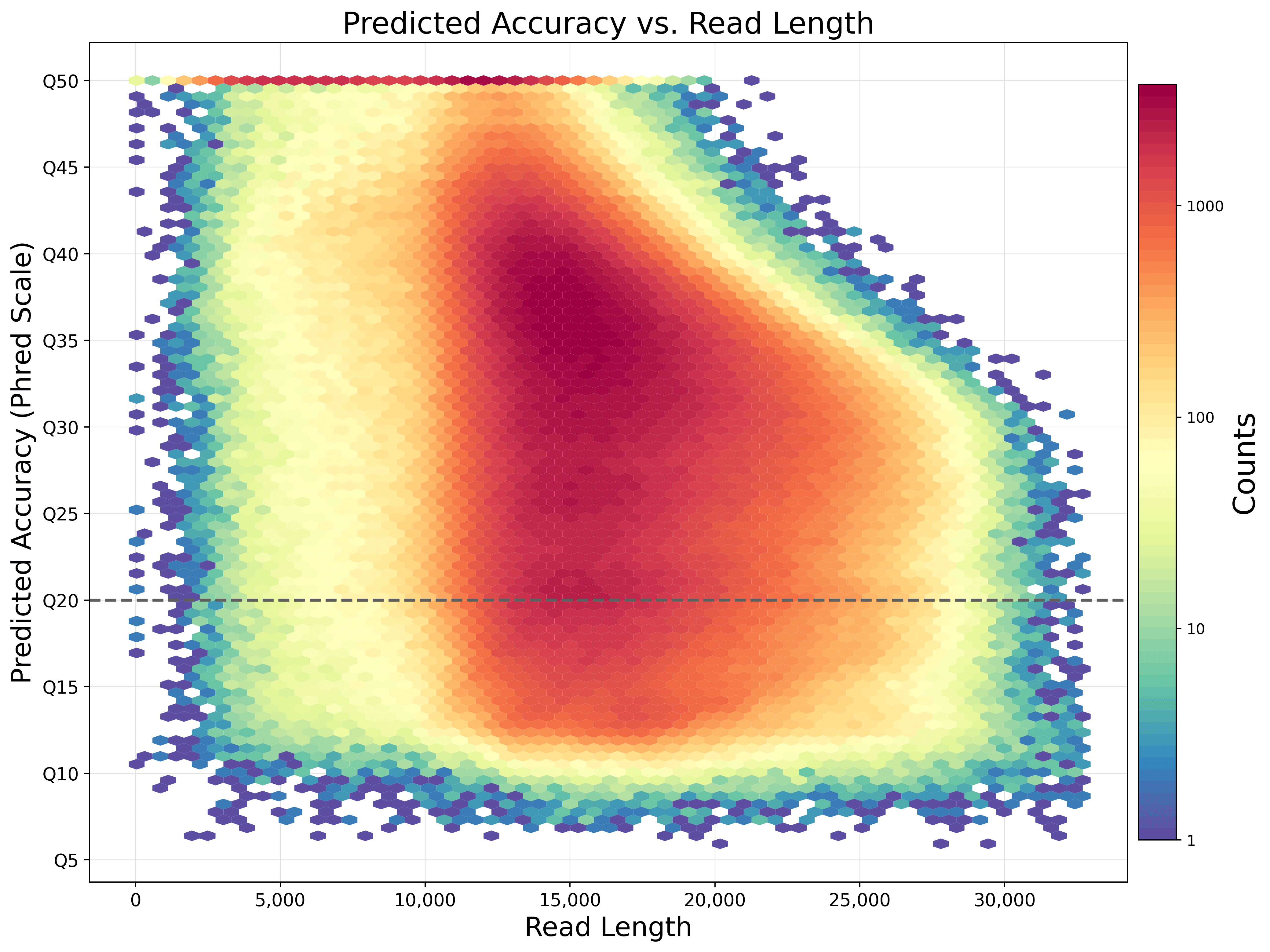
From kilobases to "whales": a short history of ultra-long reads and high-throughput genome sequencing
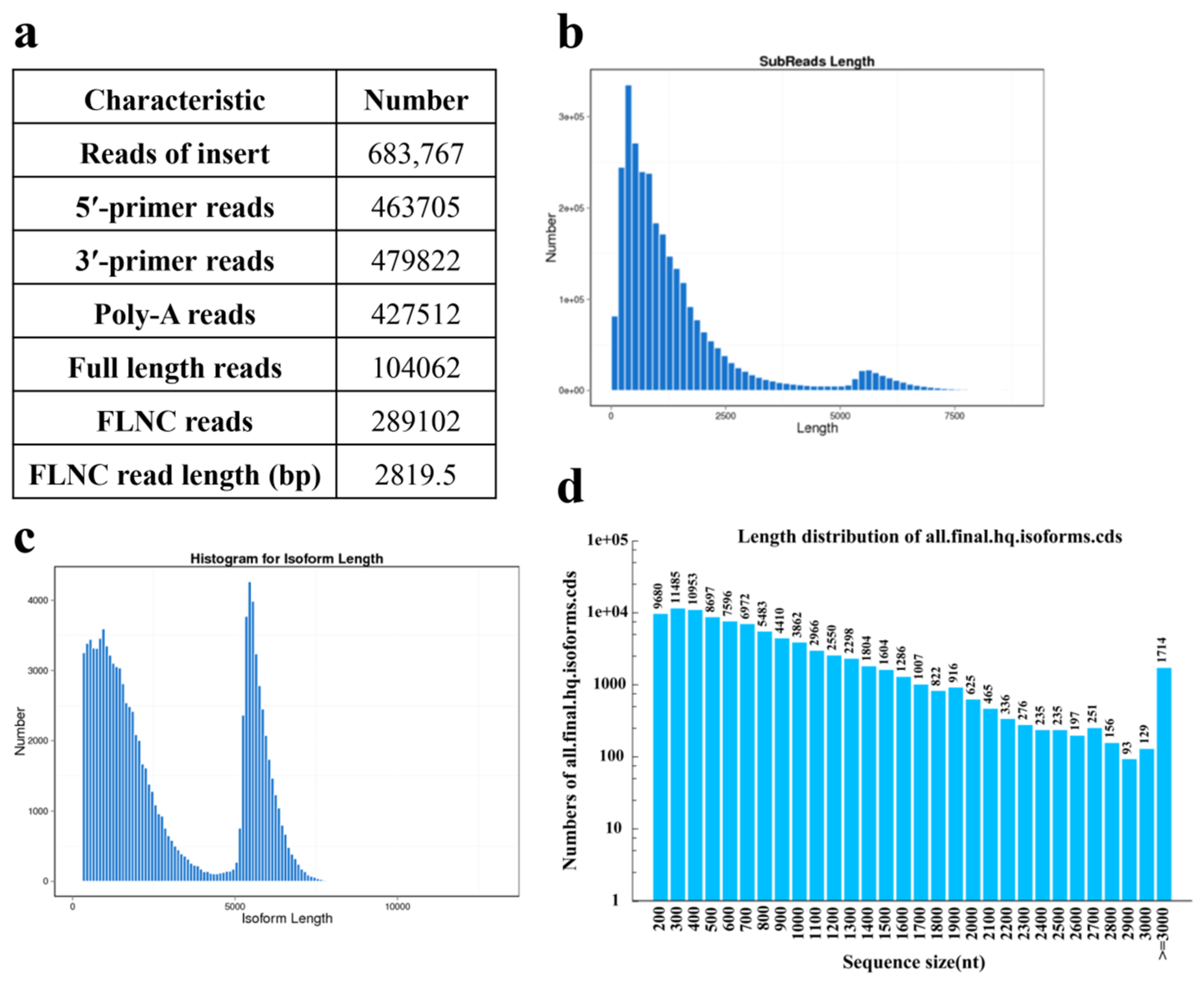
IJMS | Free Full-Text | Full-Length Transcriptome Sequencing and Different Chemotype Expression Profile Analysis of Genes Related to Monoterpenoid Biosynthesis in Cinnamomum porrectum
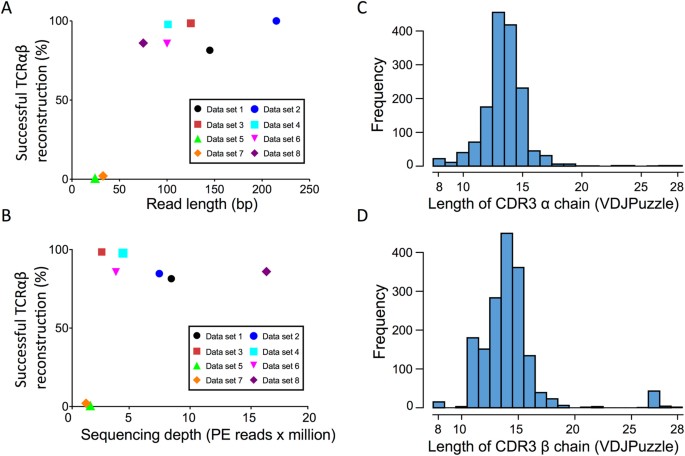
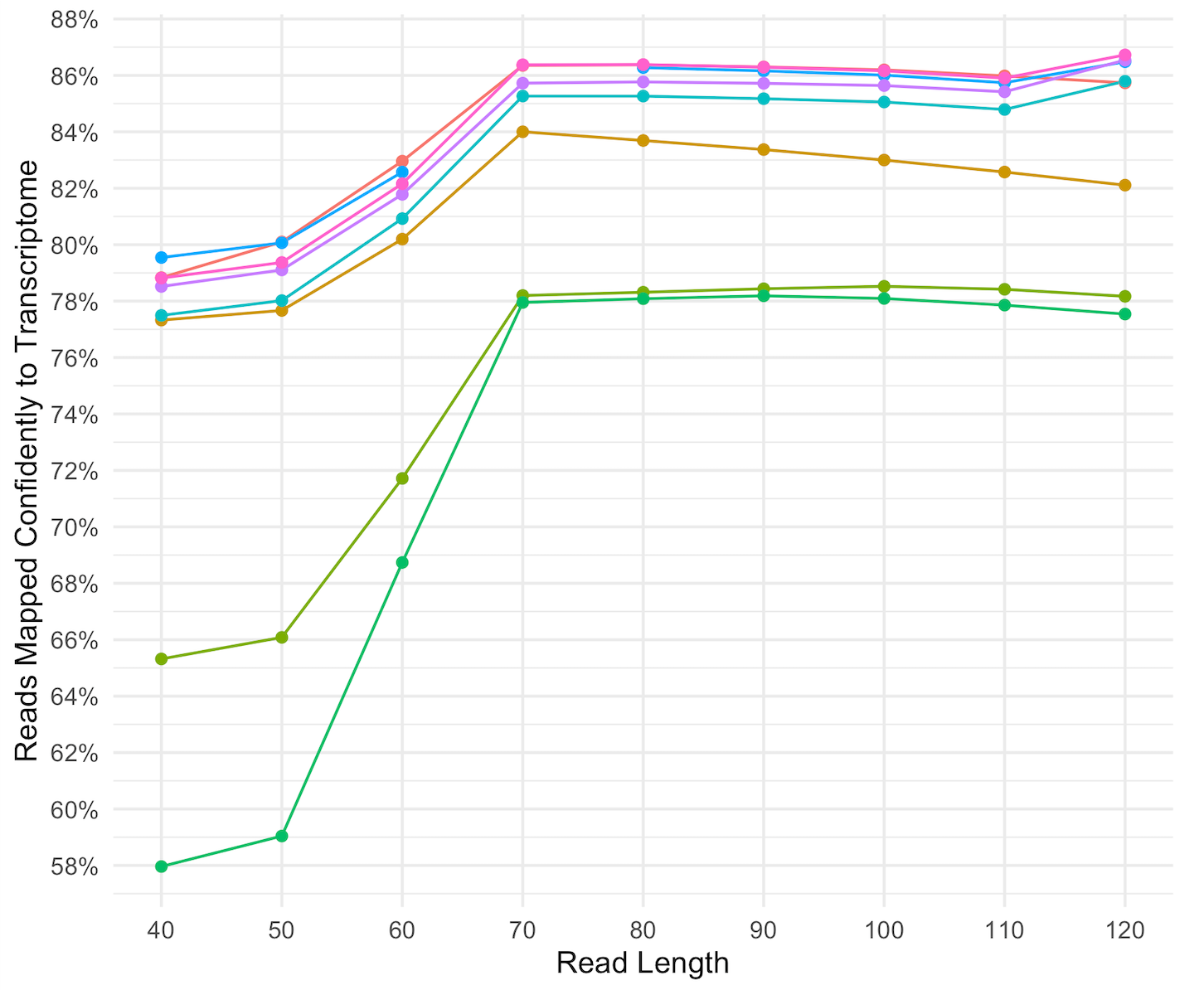
Impact of sequencing depth and read length on single cell RNA sequencing data of T cells | Scientific Reports
Direct Chloroplast Sequencing: Comparison of Sequencing Platforms and Analysis Tools for Whole Chloroplast Barcoding | PLOS ONE