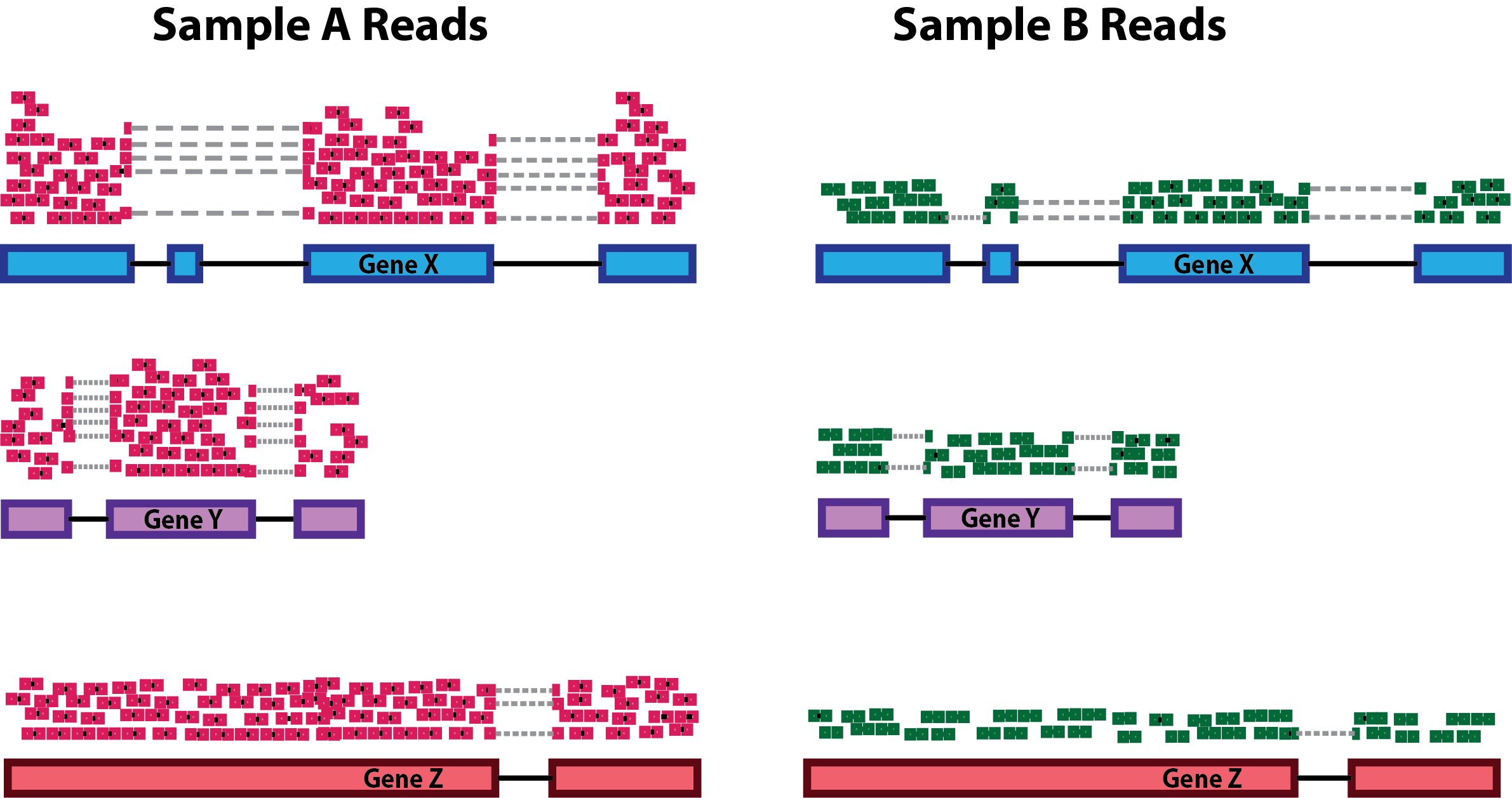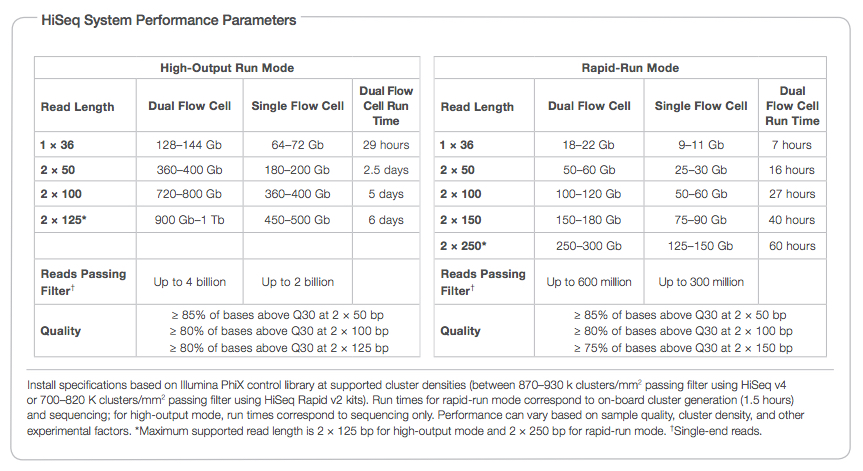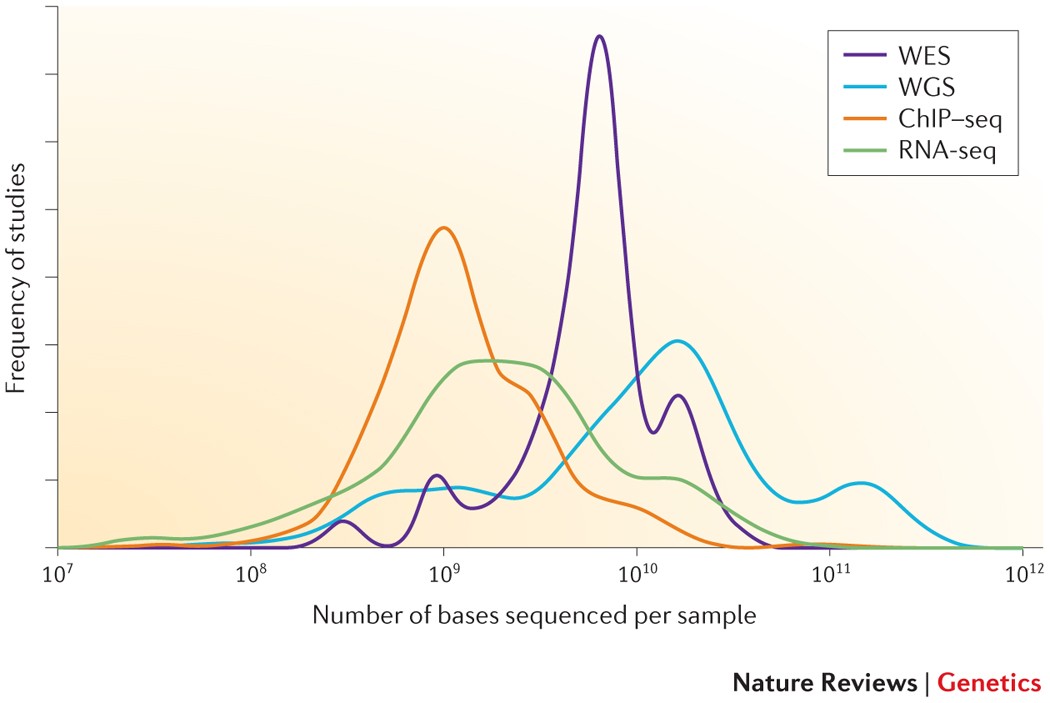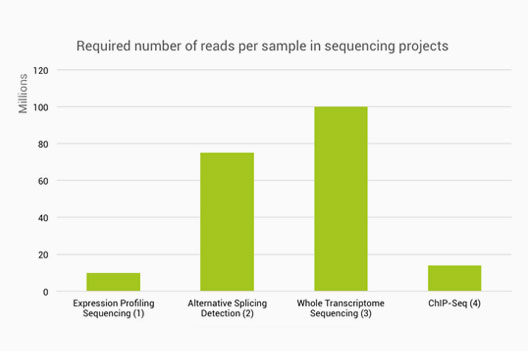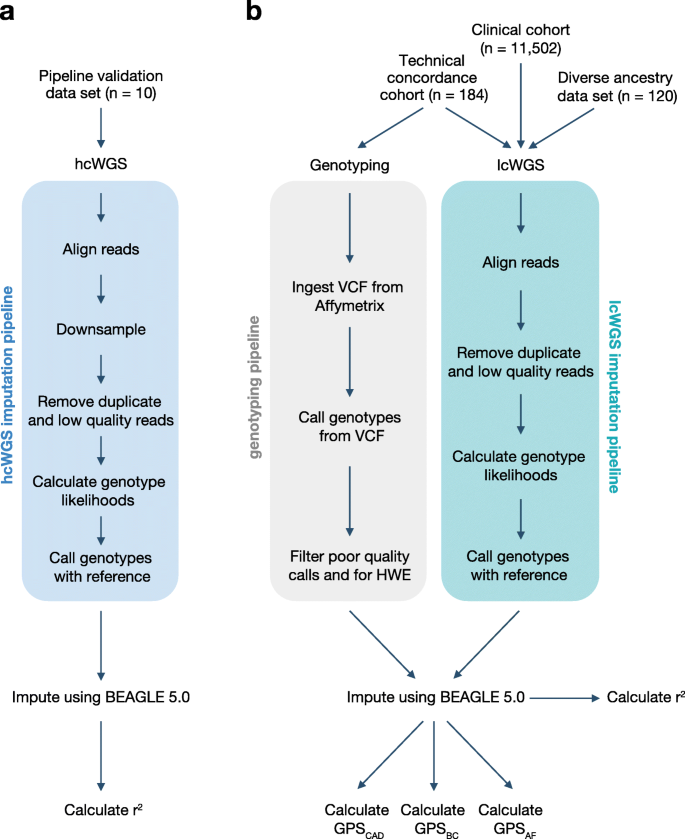
Low coverage whole genome sequencing enables accurate assessment of common variants and calculation of genome-wide polygenic scores | Genome Medicine | Full Text

A beginner's guide to low‐coverage whole genome sequencing for population genomics - Lou - 2021 - Molecular Ecology - Wiley Online Library
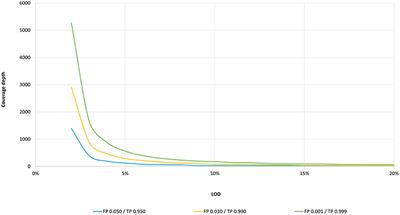
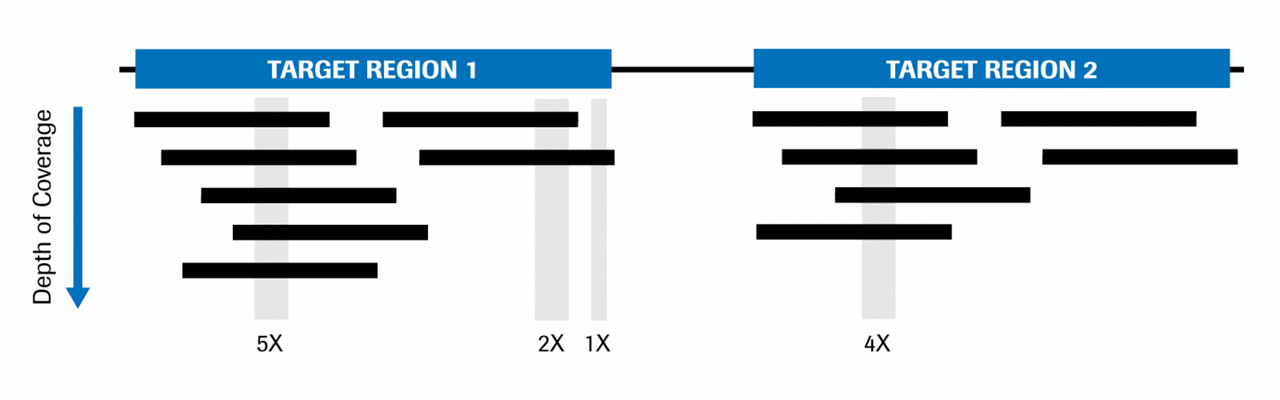
Frontiers | Standardization of Sequencing Coverage Depth in NGS: Recommendation for Detection of Clonal and Subclonal Mutations in Cancer Diagnostics
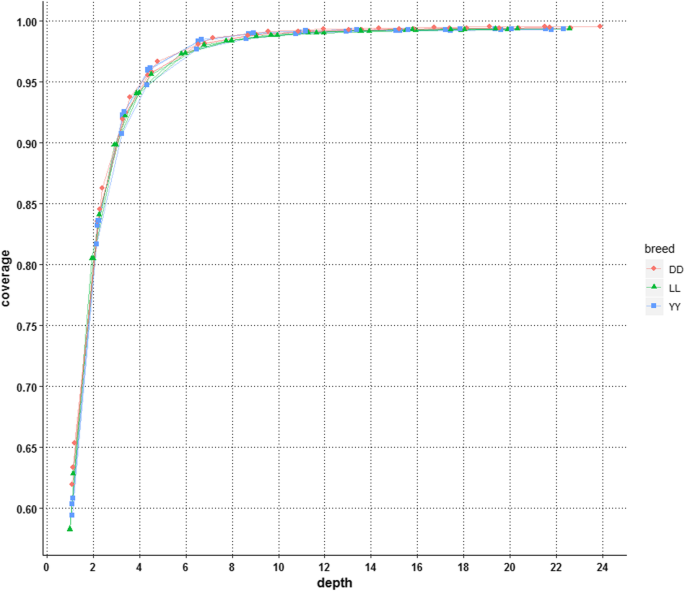
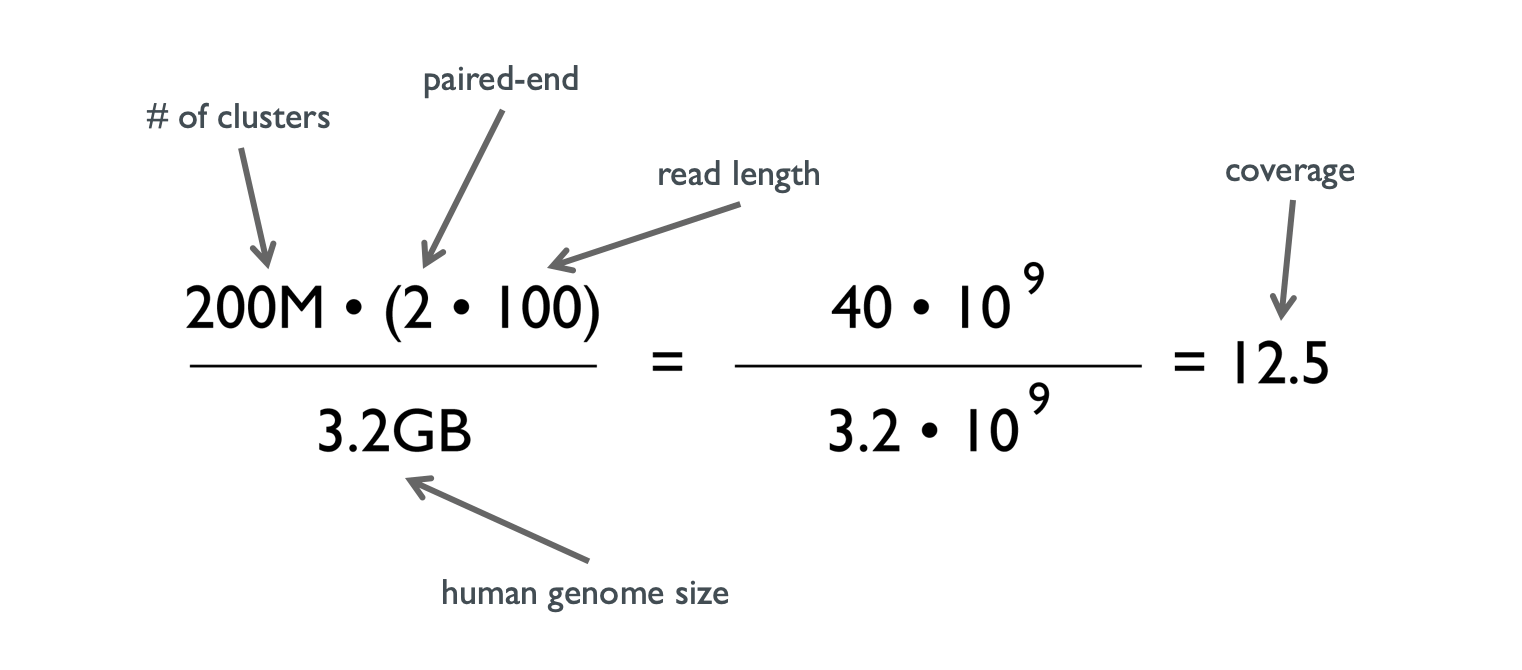
Optimal sequencing depth design for whole genome re-sequencing in pigs | BMC Bioinformatics | Full Text
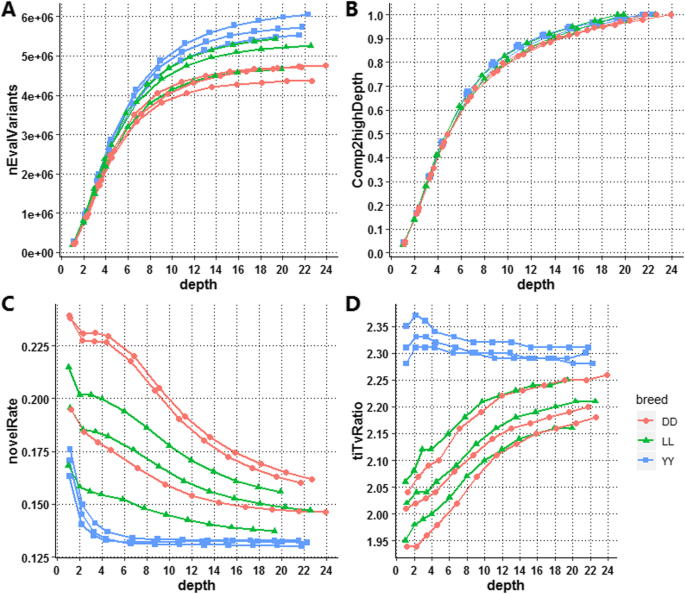
Optimal sequencing depth design for whole genome re-sequencing in pigs | BMC Bioinformatics | Full Text

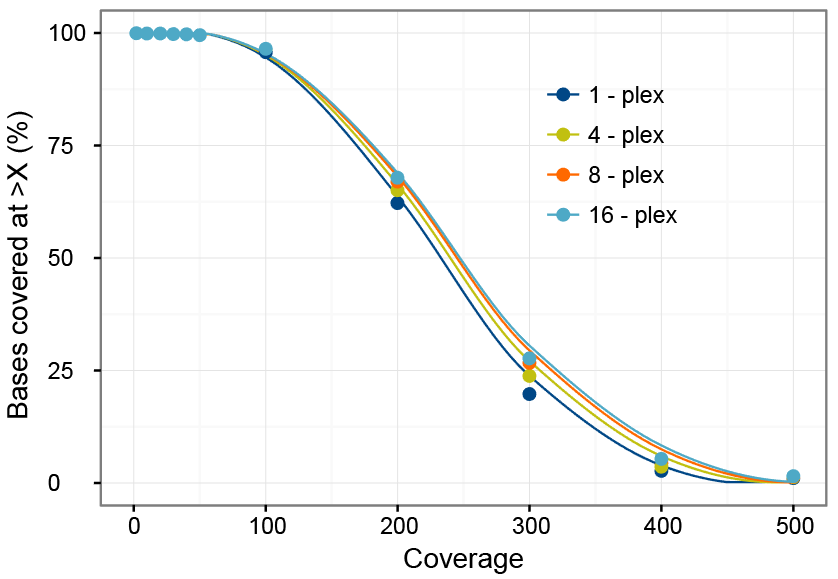
Mathematical Framework Provides a Read Depth Calculator and Guidelines... | Download Scientific Diagram