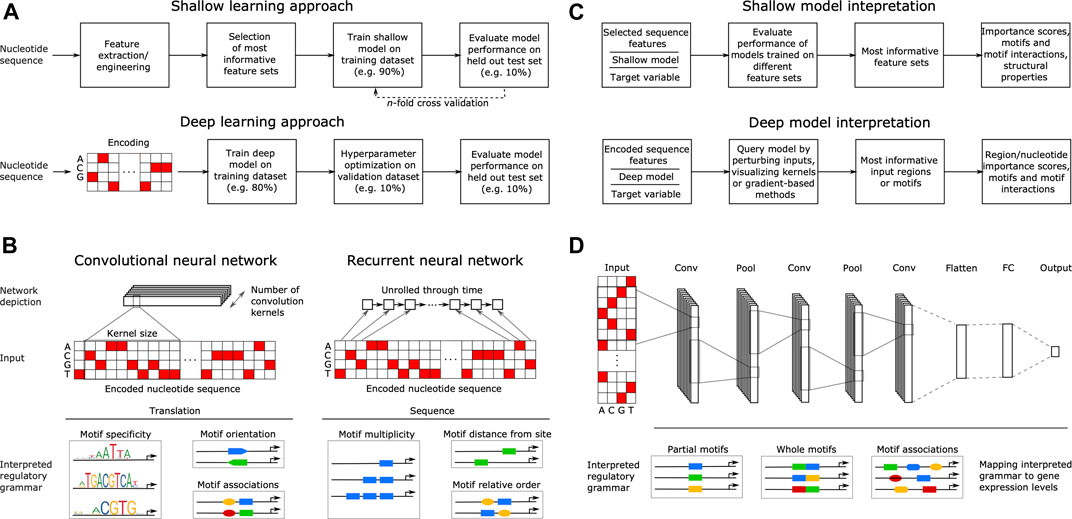
Machine learning predicts translation initiation sites in neurologic diseases with expanded repeats | bioRxiv
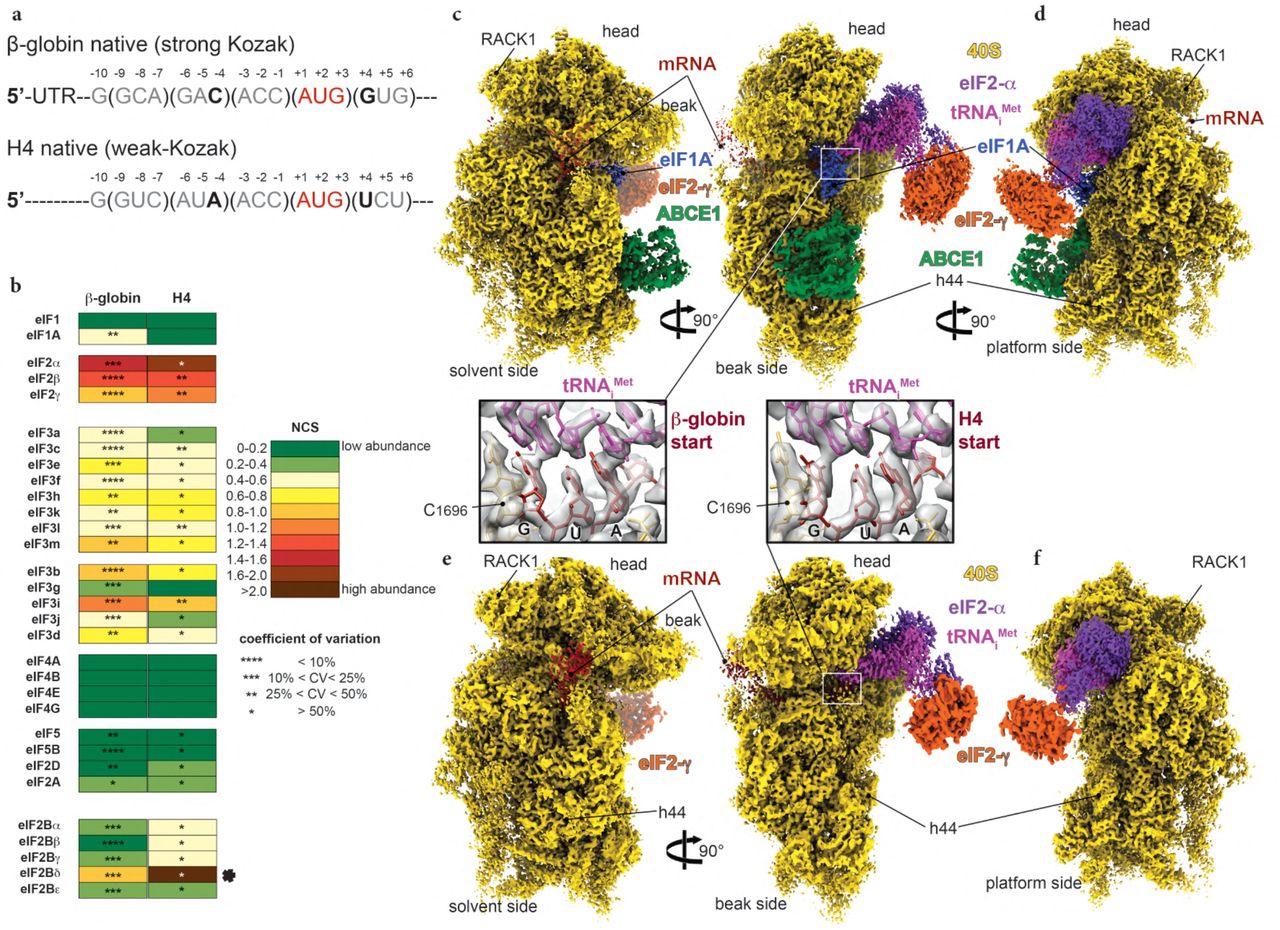
Structural insights into the Kozak sequence interactions within the mammalian late-stage initiation complexes | bioRxiv

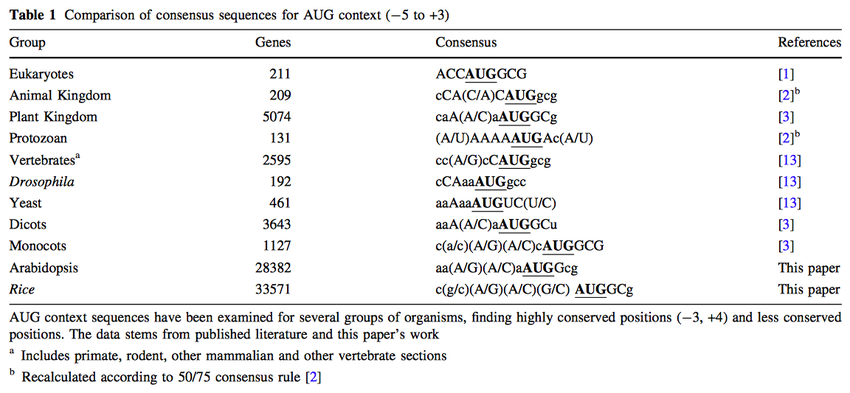
Bioinformatic analyses of mammalian 5'-UTR sequence properties of mRNAs predicts alternative translation initiation sites | BMC Bioinformatics | Full Text
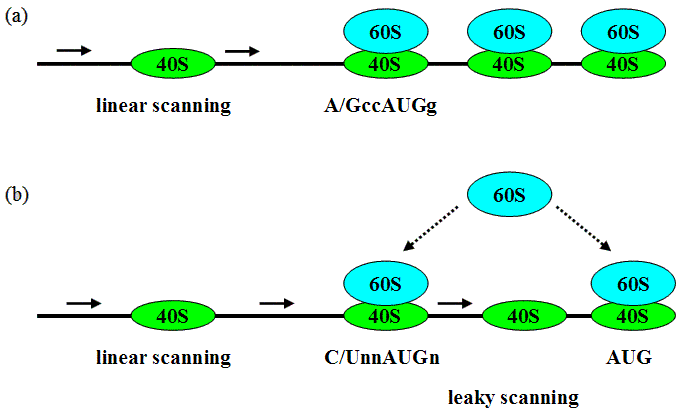
AUG_hairpin: program for prediction of a downstream hairpin potentially increasing initiation of translation at start AUG codon in a suboptimal context.

Structural insights into the Kozak sequence interactions within the mammalian late-stage initiation complexes | bioRxiv

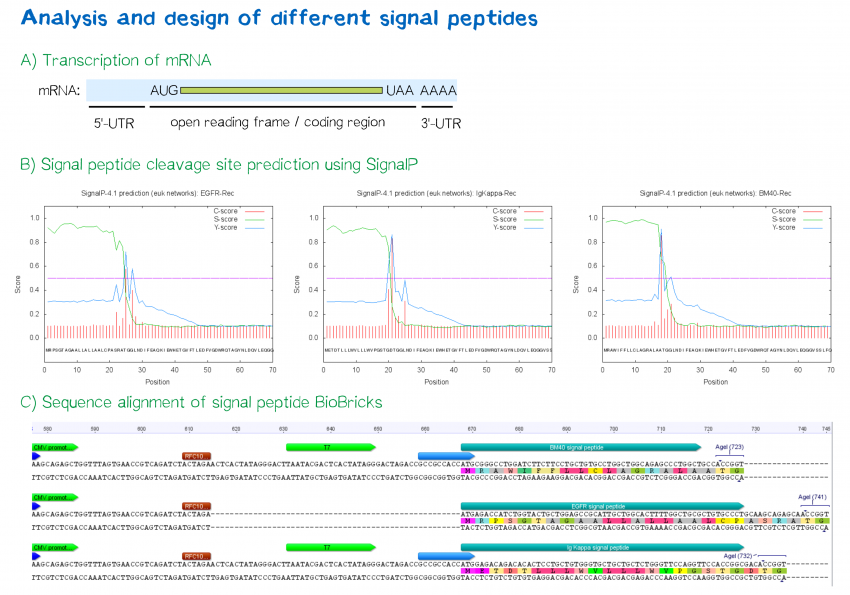
Vaccines | Free Full-Text | mRNA Vaccine Designing Using Chikungunya Virus E Glycoprotein through Immunoinformatics-Guided Approaches
The +4G Site in Kozak Consensus Is Not Related to the Efficiency of Translation Initiation | PLOS ONE

Quantitative analysis of mammalian translation initiation sites by FACS‐seq | Molecular Systems Biology
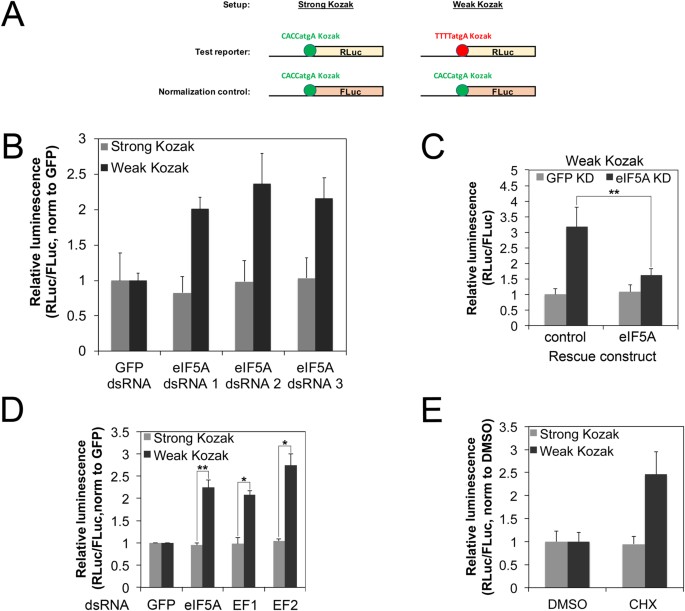
Changes in global translation elongation or initiation rates shape the proteome via the Kozak sequence | Scientific Reports

Quantification and discovery of sequence determinants of protein‐per‐mRNA amount in 29 human tissues | Molecular Systems Biology

The effect of changing the +4 position of the Kozak sequence on the... | Download Scientific Diagram
Machine learning predicts translation initiation sites in neurologic diseases with nucleotide repeat expansions | PLOS ONE

Rps26 directs mRNA-specific translation by recognition of Kozak sequence elements | Nature Structural & Molecular Biology

DeepTIS: Improved translation initiation site prediction in genomic sequence via a two-stage deep learning model - ScienceDirect