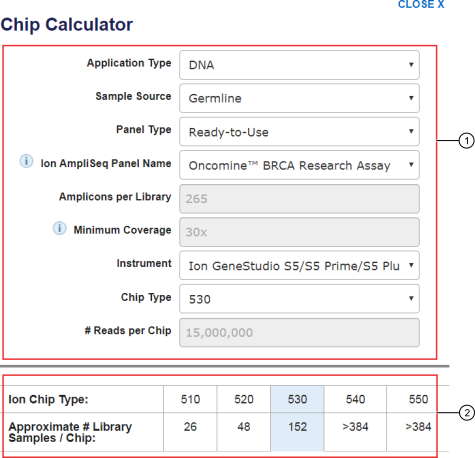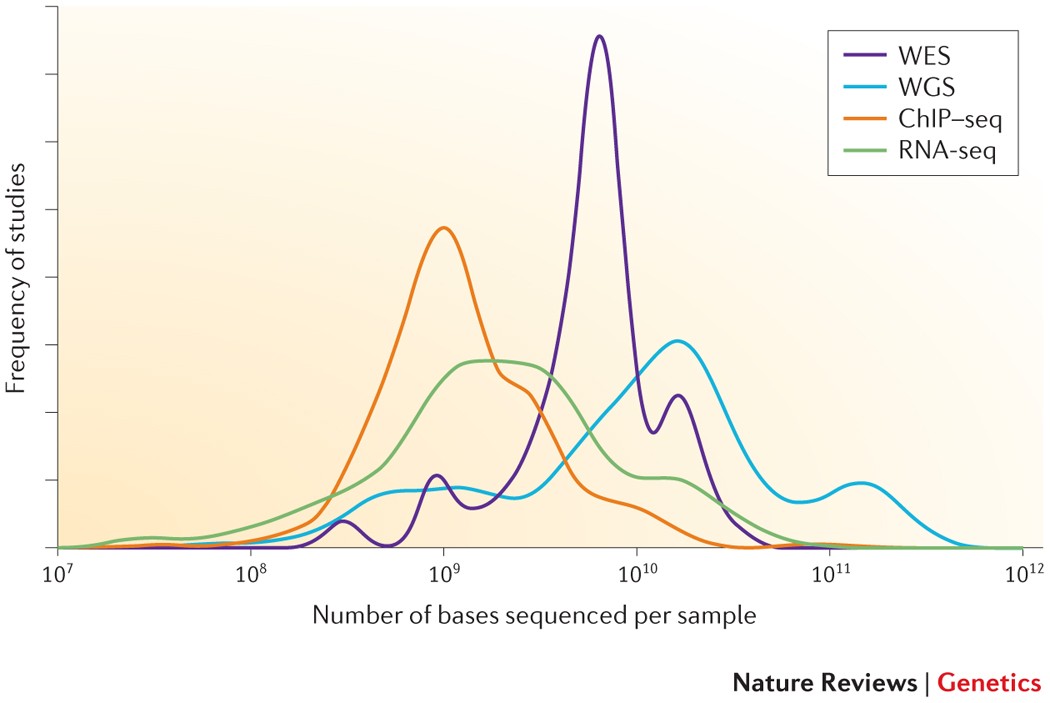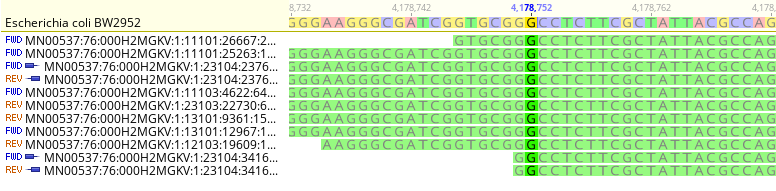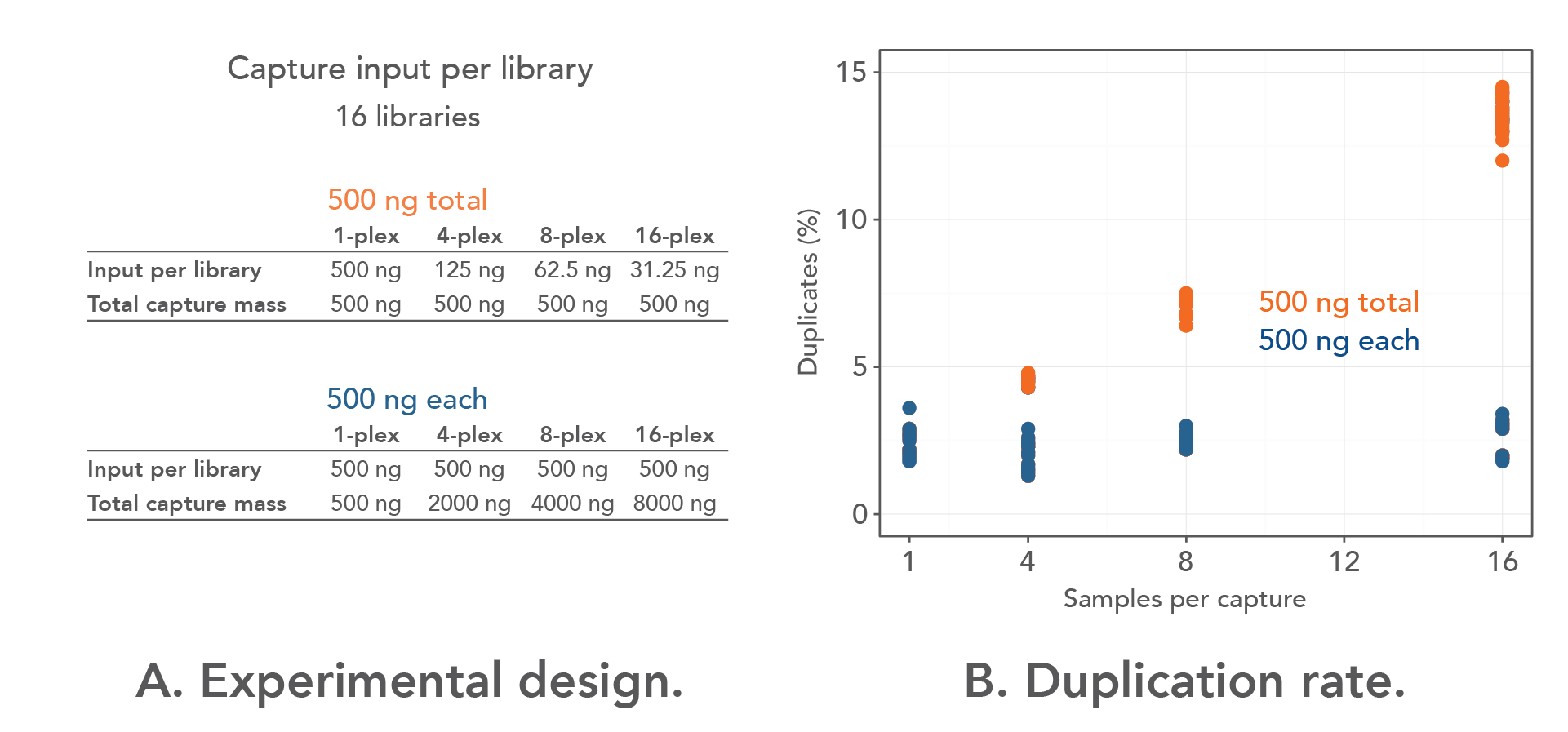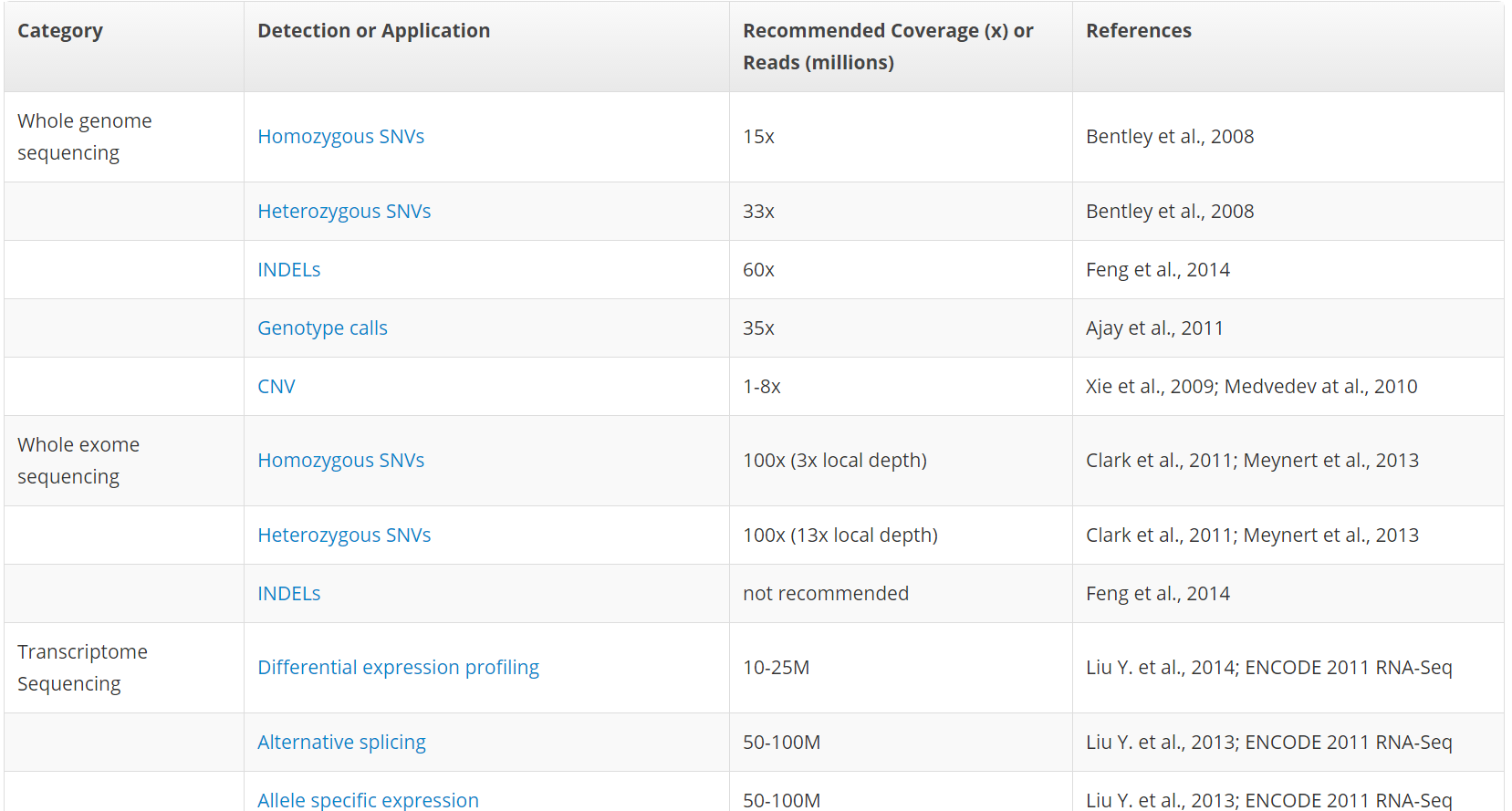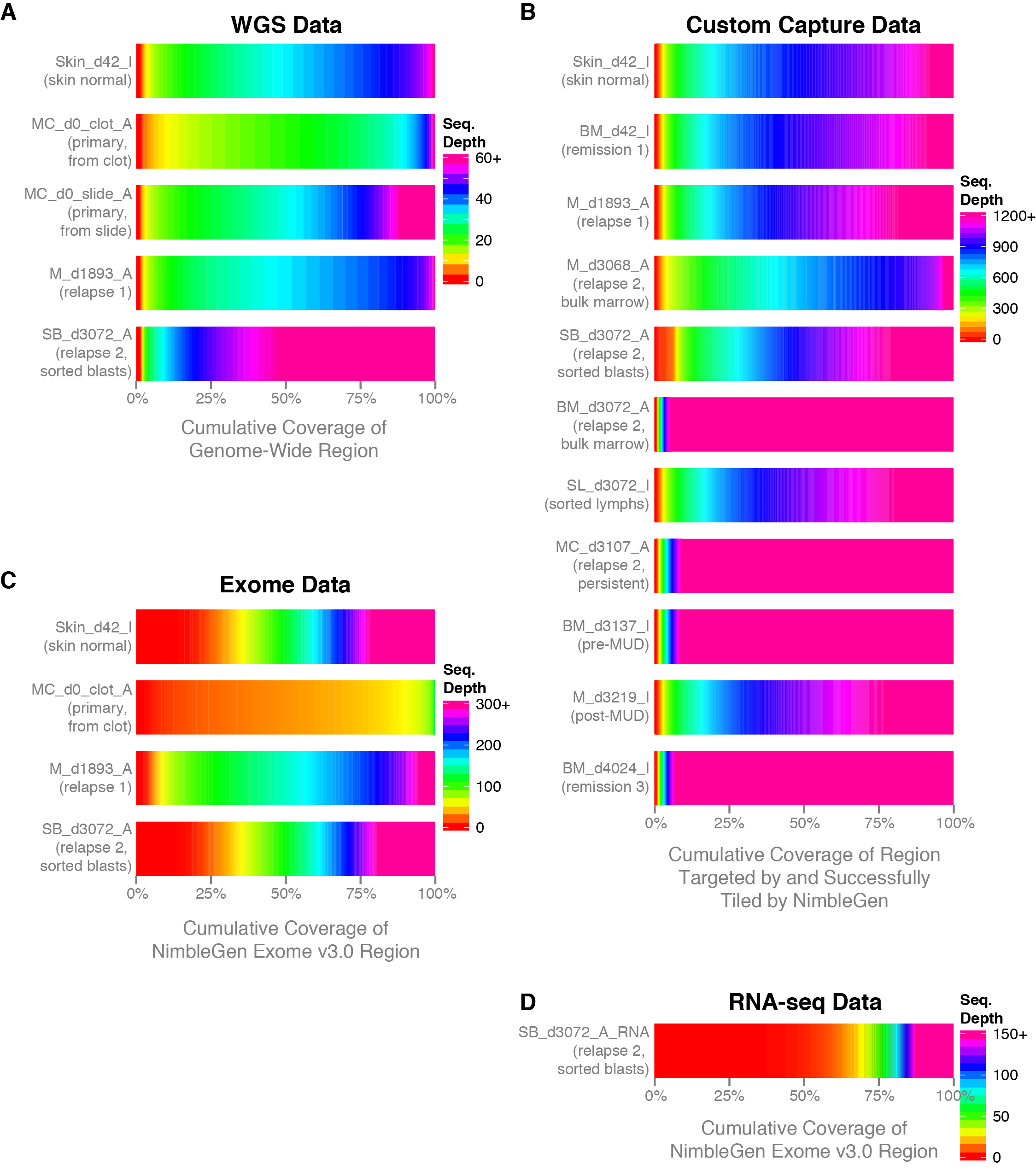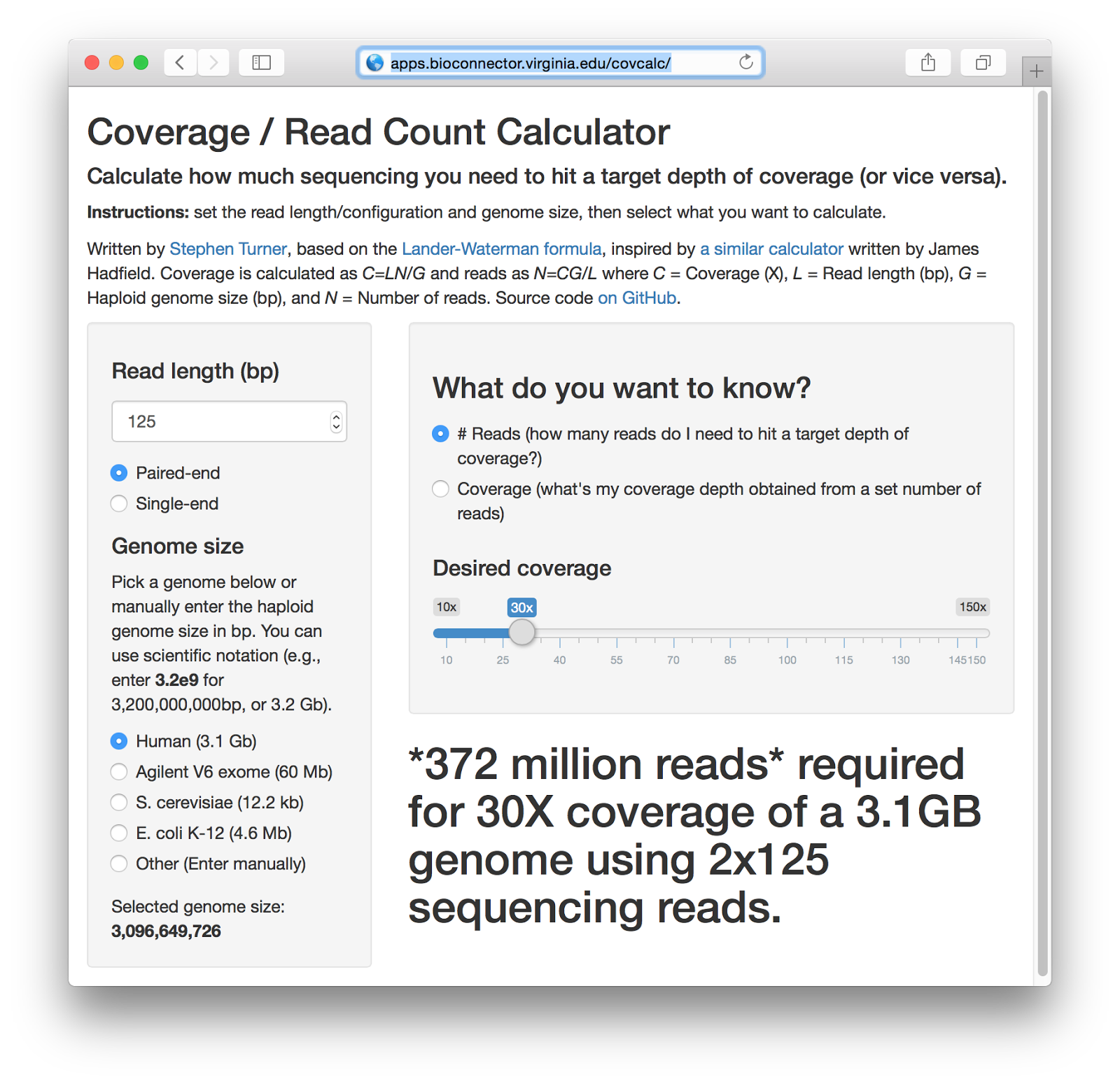
Getting Genetics Done: Covcalc: Shiny App for Calculating Coverage Depth or Read Counts for Sequencing Experiments
PDF) Standardization of Sequencing Coverage Depth in NGS: Recommendation for Detection of Clonal and Subclonal Mutations in Cancer Diagnostics
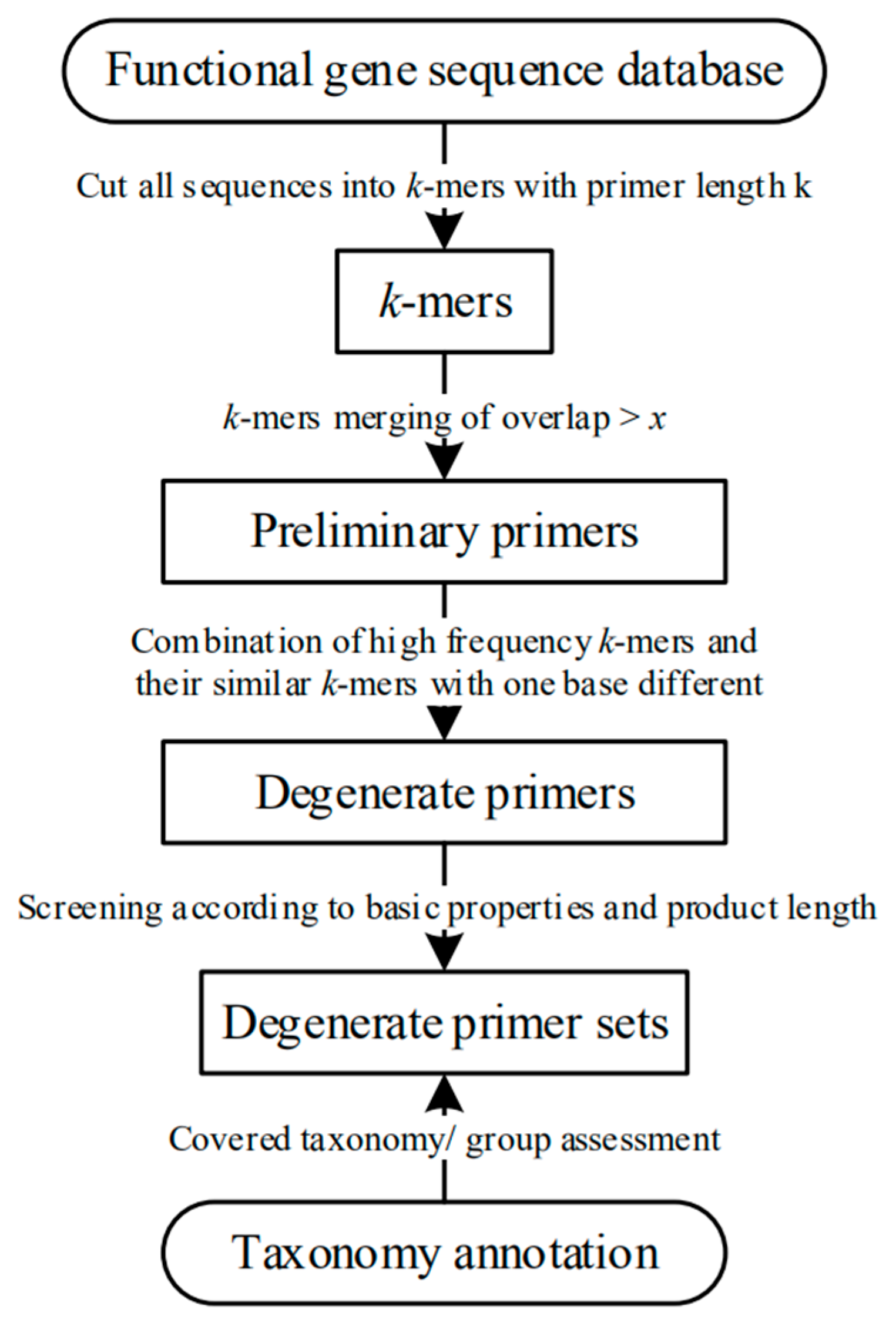
IJERPH | Free Full-Text | ARDEP, a Rapid Degenerate Primer Design Pipeline Based on k-mers for Amplicon Microbiome Studies
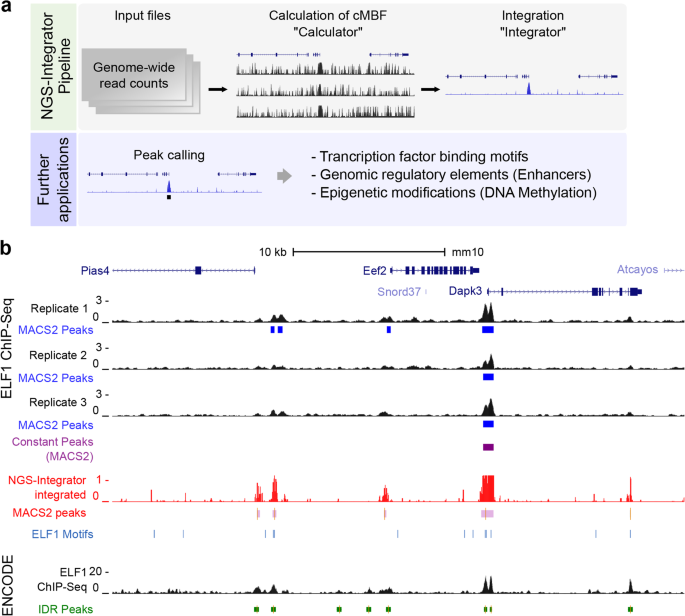
NGS-Integrator: An efficient tool for combining multiple NGS data tracks using minimum Bayes' factors | BMC Genomics | Full Text
CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing | PLOS Computational Biology

Devyser on Twitter: "Did you know we have a Coverage Calculator which can help your sequencing planning? Just select your system and kit, the number and type of samples, and easily calculate

How is the percentage of protein sequence coverage calculated in the search report of MS/MS Ions Search in Mascot? | ResearchGate
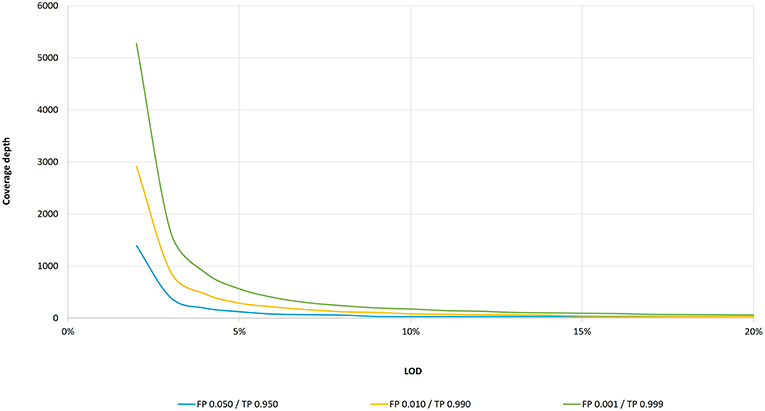
Frontiers | Standardization of Sequencing Coverage Depth in NGS: Recommendation for Detection of Clonal and Subclonal Mutations in Cancer Diagnostics