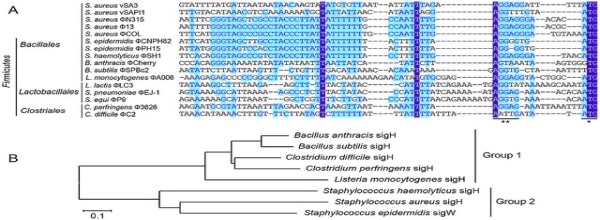
Example of pseudomultiple sequence alignment from PSI-Blast.A cropped... | Download Scientific Diagram

by sequence - NCBI Resources - LibGuides at Health Sciences Library, University of Colorado Anschutz Medical Campus

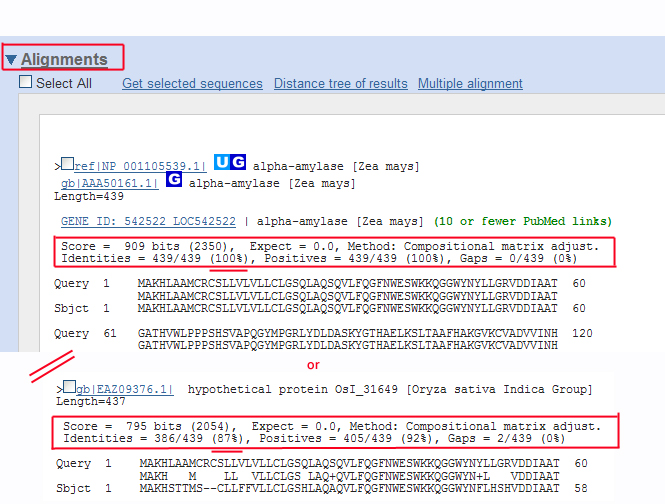
The protein BLAST (BLASTp) search against the annotated NCBI protein... | Download Scientific Diagram

NCBI Bioinformatics Tools: Protein, BLAST, COBALT, and Cn3D Structure Viewer – Inside Science Resources
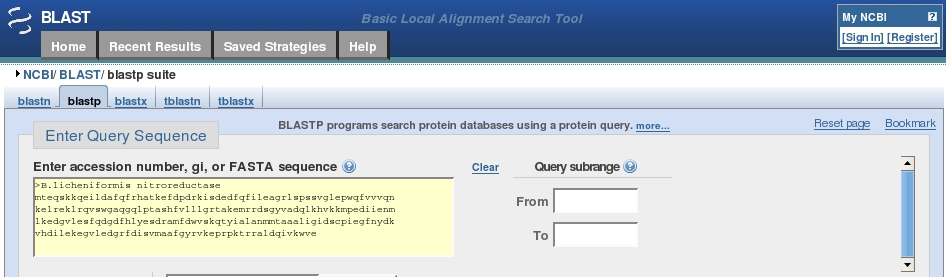
Protein BLAST allows one to input protein sequences and compare these against other protein sequences
![PDF] BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. | Semantic Scholar PDF] BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/92dffee93794394dd7d1b2db3dcb69177e0560cb/3-Figure1-1.png)
PDF] BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. | Semantic Scholar

What is BLAST? BLAST® (Basic Local Alignment Search Tool) is a set of similarity search programs designed to explore all of the available sequence databases. - ppt video online download

BLAST: Compare & identify sequences - NCBI Bioinformatics Resources: An Introduction - Library Guides at UC Berkeley
NCBI Bioinformatics Tools: Protein, BLAST, COBALT, and Cn3D Structure Viewer – Inside Science Resources

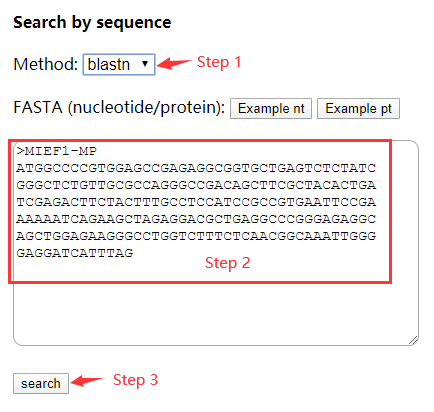
BLASTP Protein Sequence Search - Introduction to NCBI Bioinformatics Resources - Learning Resource Center at Uniformed Services University of the Health Sciences