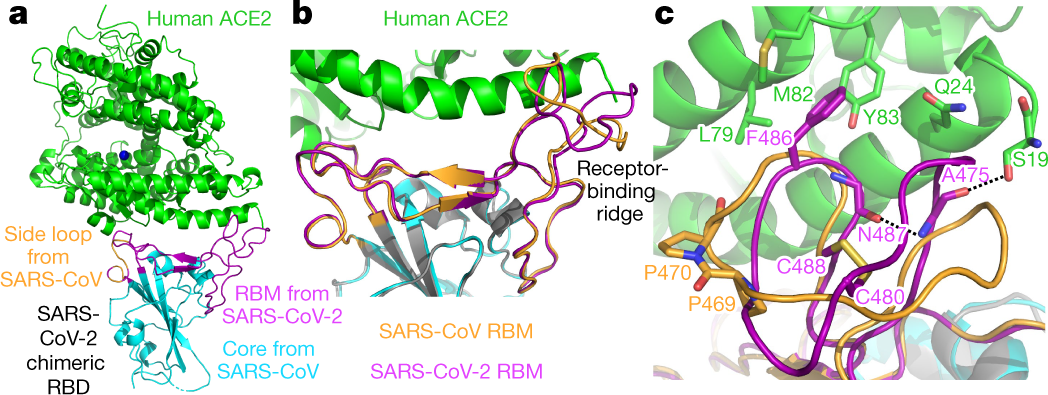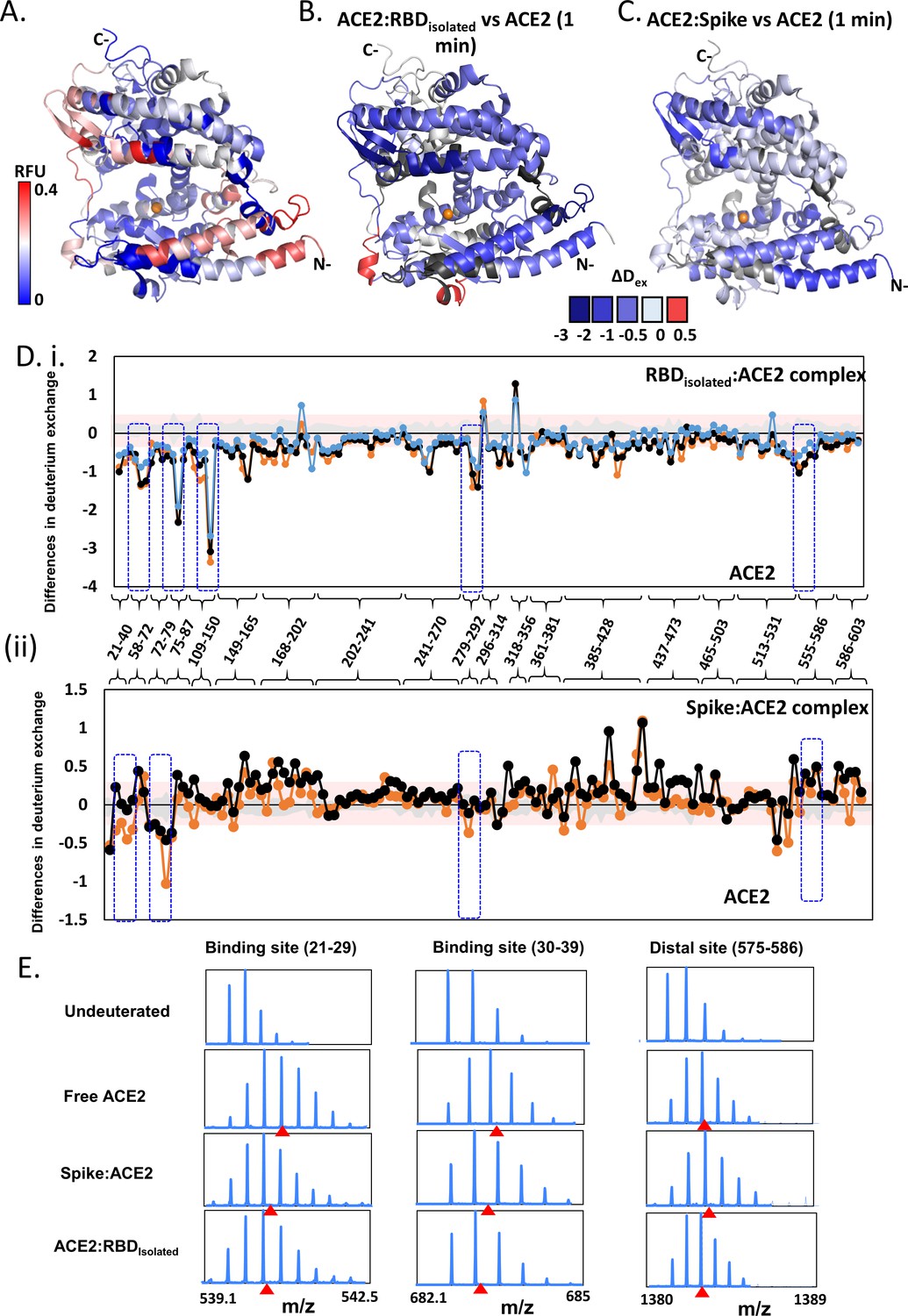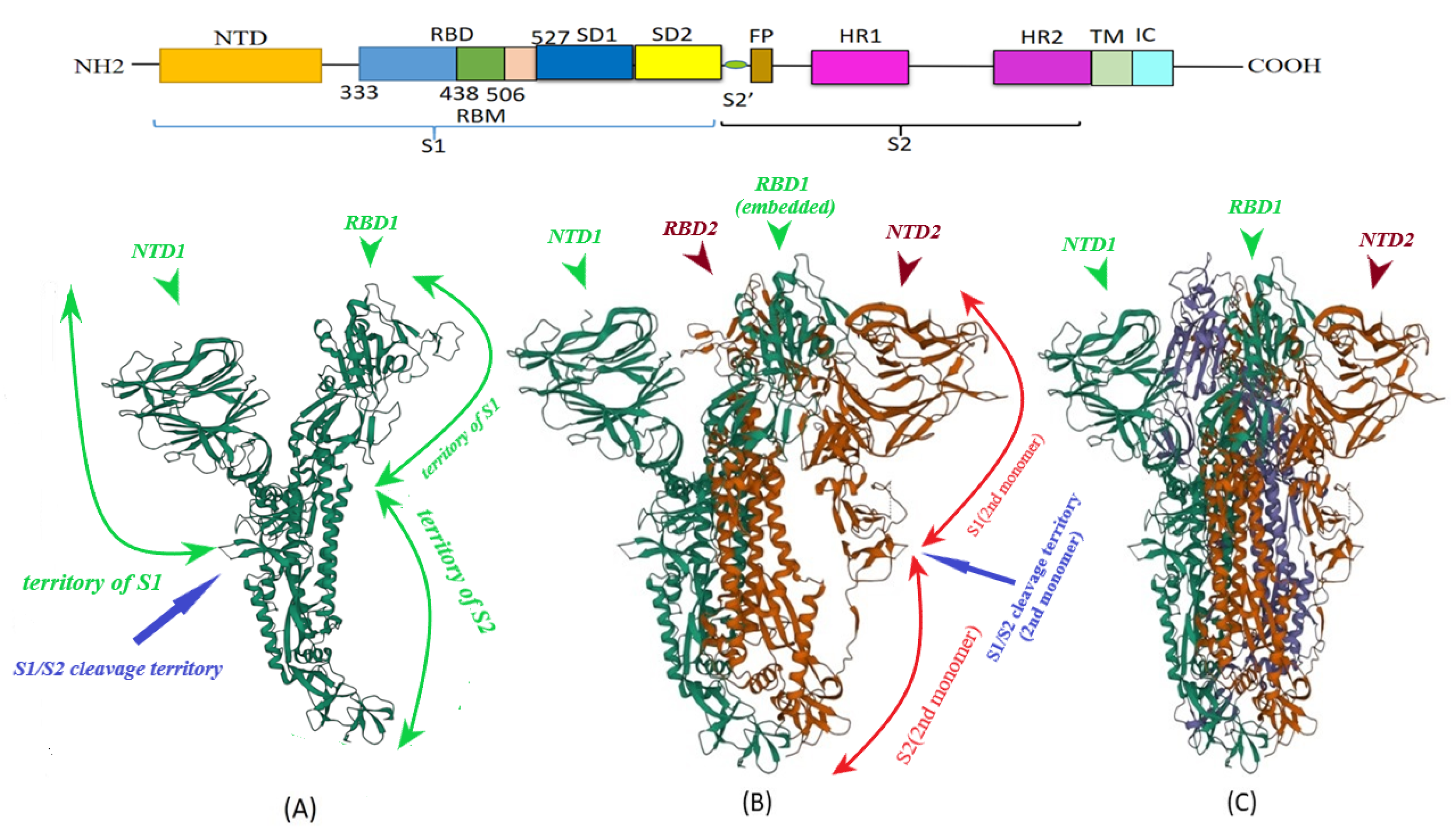
Vaccines | Free Full-Text | S Protein, ACE2 and Host Cell Proteases in SARS-CoV-2 Cell Entry and Infectivity; Is Soluble ACE2 a Two Blade Sword? A Narrative Review
Discovery of human ACE2 variants with altered recognition by the SARS-CoV-2 spike protein | PLOS ONE

Synthetic α‐Helical Peptides as Potential Inhibitors of the ACE2 SARS‐CoV‐2 Interaction - Engelhardt - 2022 - ChemBioChem - Wiley Online Library

Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor | Nature Communications
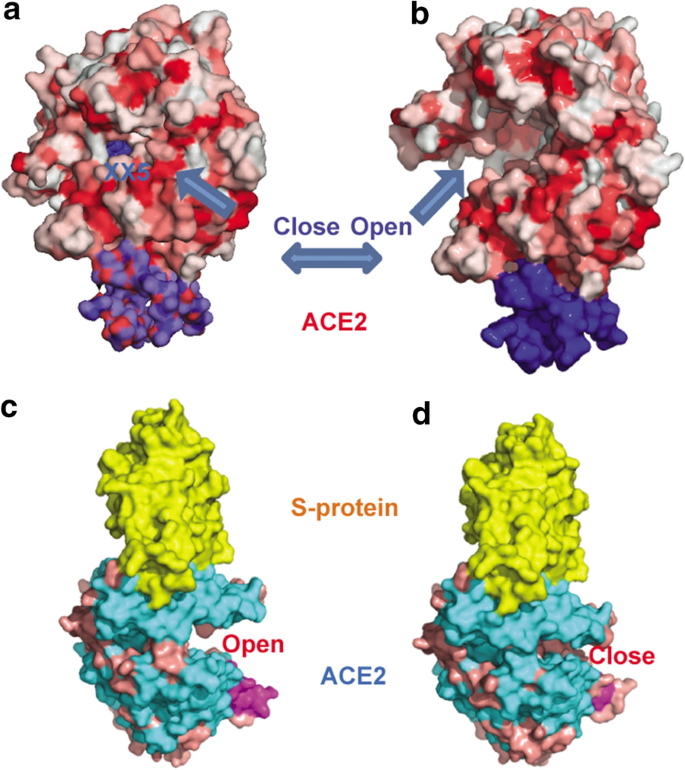
Investigation of the genetic variation in ACE2 on the structural recognition by the novel coronavirus (SARS-CoV-2) | Journal of Translational Medicine | Full Text
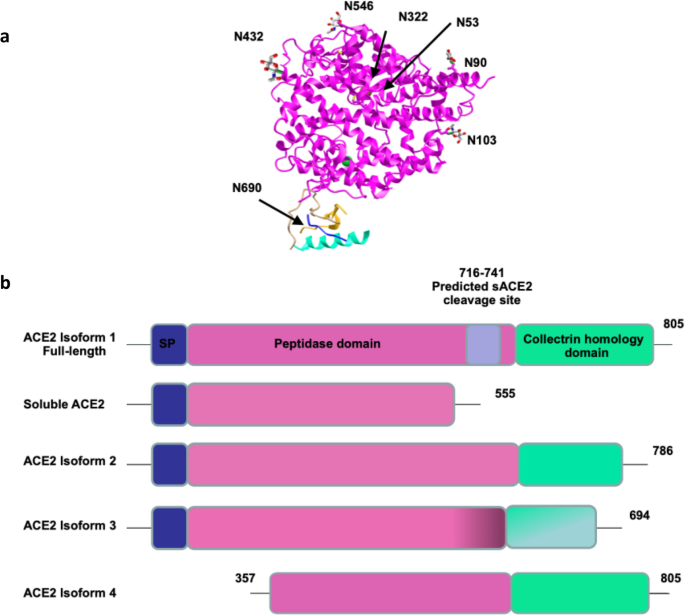
ACE2 Nascence, trafficking, and SARS-CoV-2 pathogenesis: the saga continues | Human Genomics | Full Text
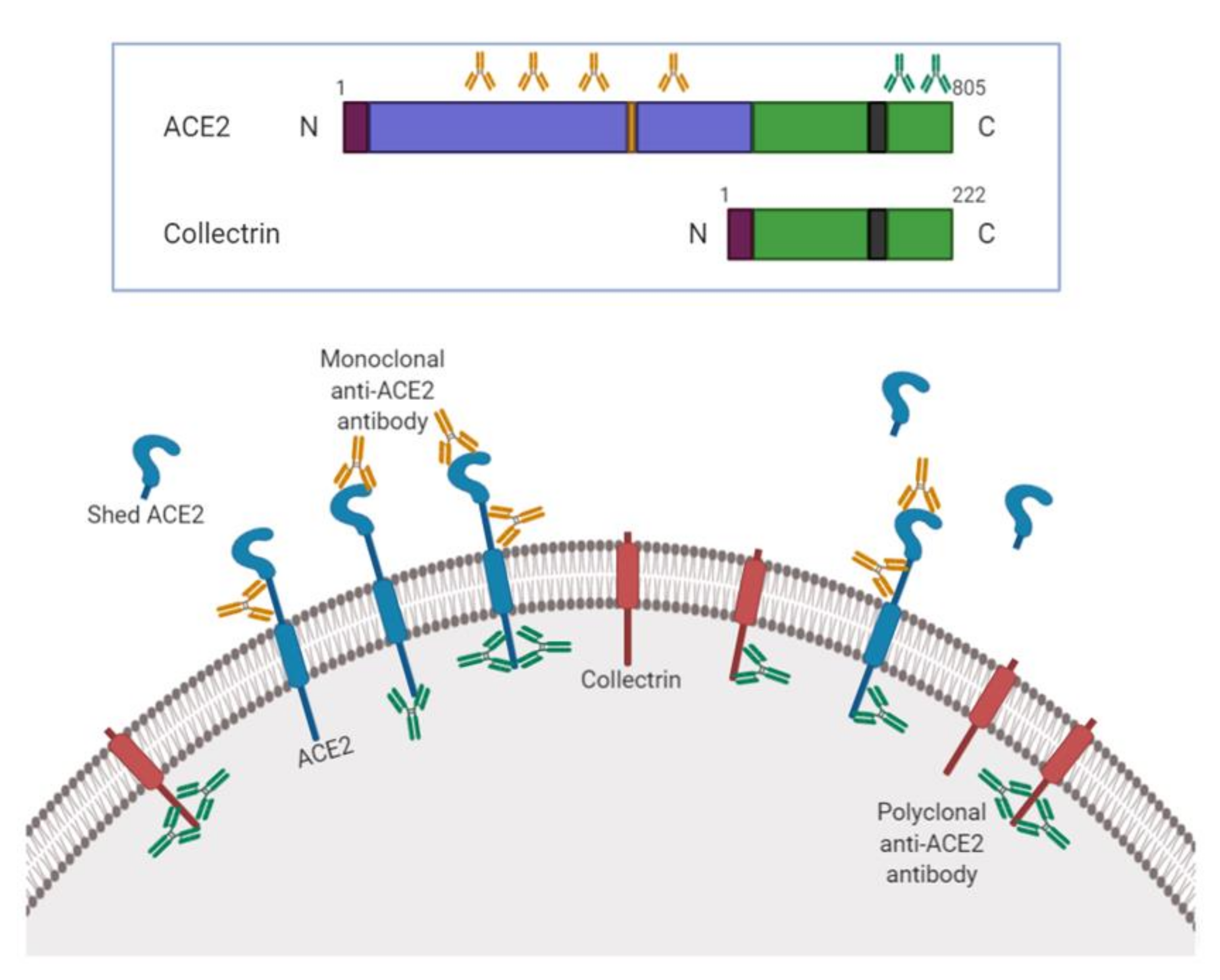
Biology | Free Full-Text | ACE2 Protein Landscape in the Head and Neck Region: The Conundrum of SARS-CoV-2 Infection
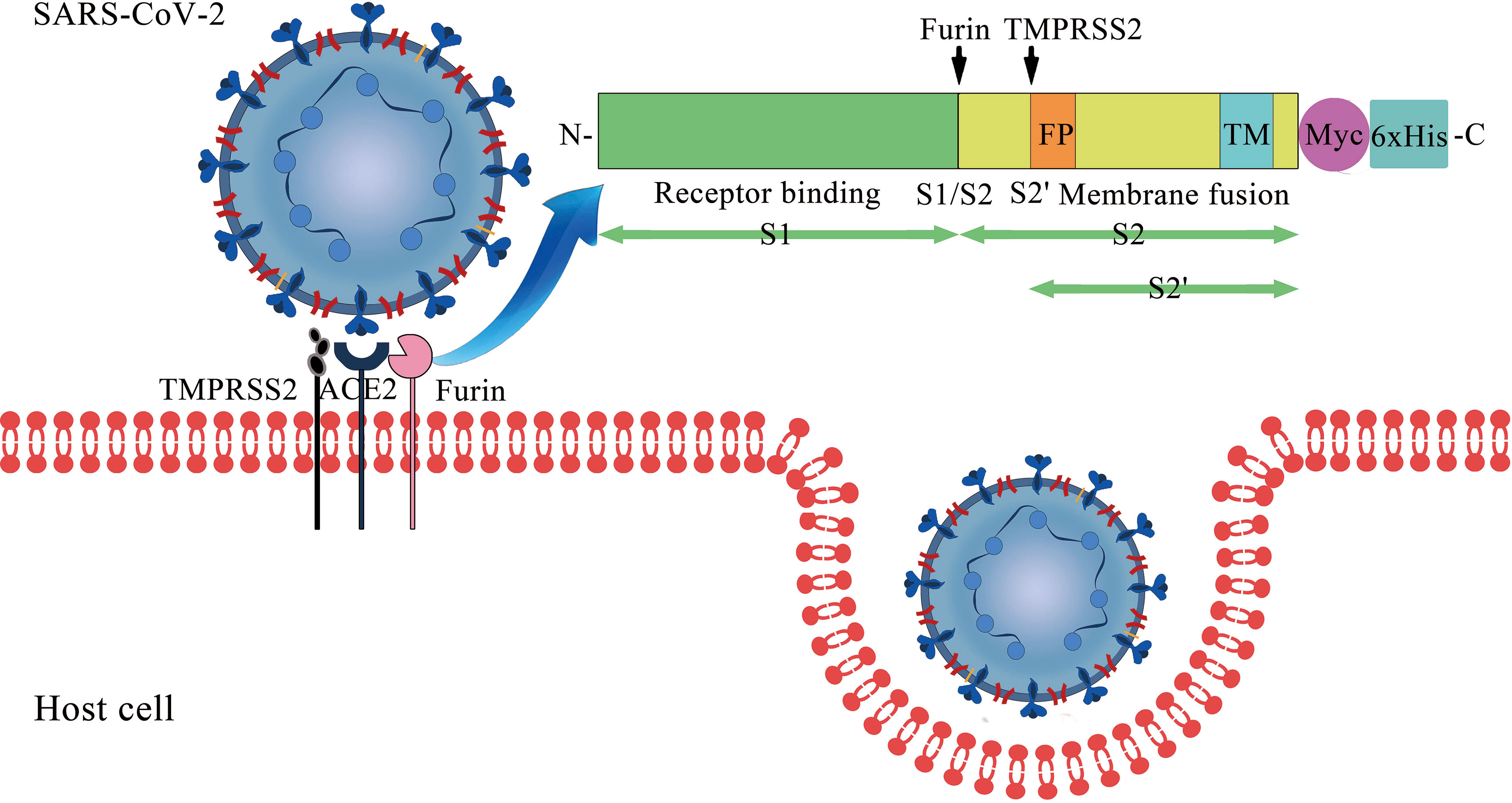
Frontiers | The Impact of ACE2 Polymorphisms on COVID-19 Disease: Susceptibility, Severity, and Therapy

An Electrostatically-steered Conformational Selection Mechanism Promotes SARS-CoV-2 Spike Protein Variation - ScienceDirect

SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor - ScienceDirect

Does SARS-CoV-2 Bind to Human ACE2 More Strongly Than Does SARS-CoV? | The Journal of Physical Chemistry B

Structural models of human ACE2 variants with SARS-CoV-2 Spike protein for structure-based drug design | Scientific Data

SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein–ACE2 complex | Science

All-Atom Simulations of Human ACE2-Spike Protein RBD Complexes for SARS-CoV-2 and Some of its Variants: Nature of Interactions and Free Energy Diagrams for Dissociation of the Protein Complexes | The Journal of