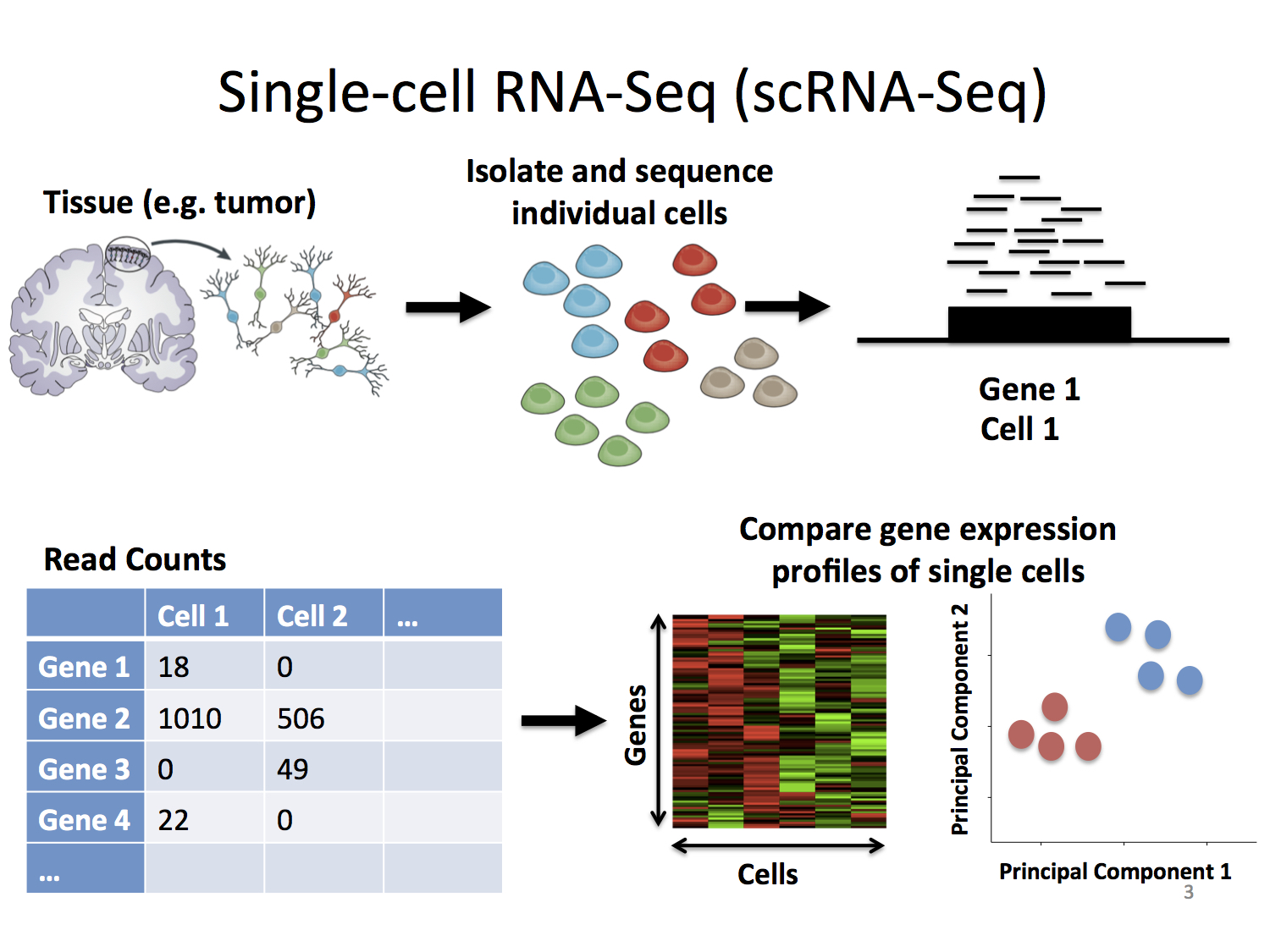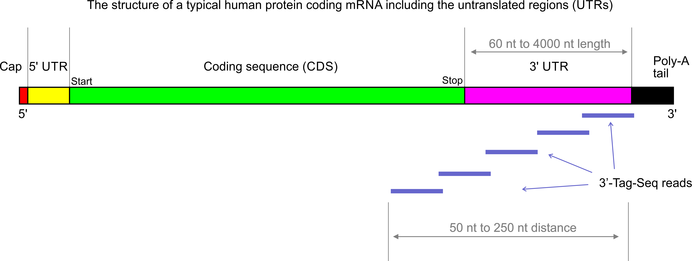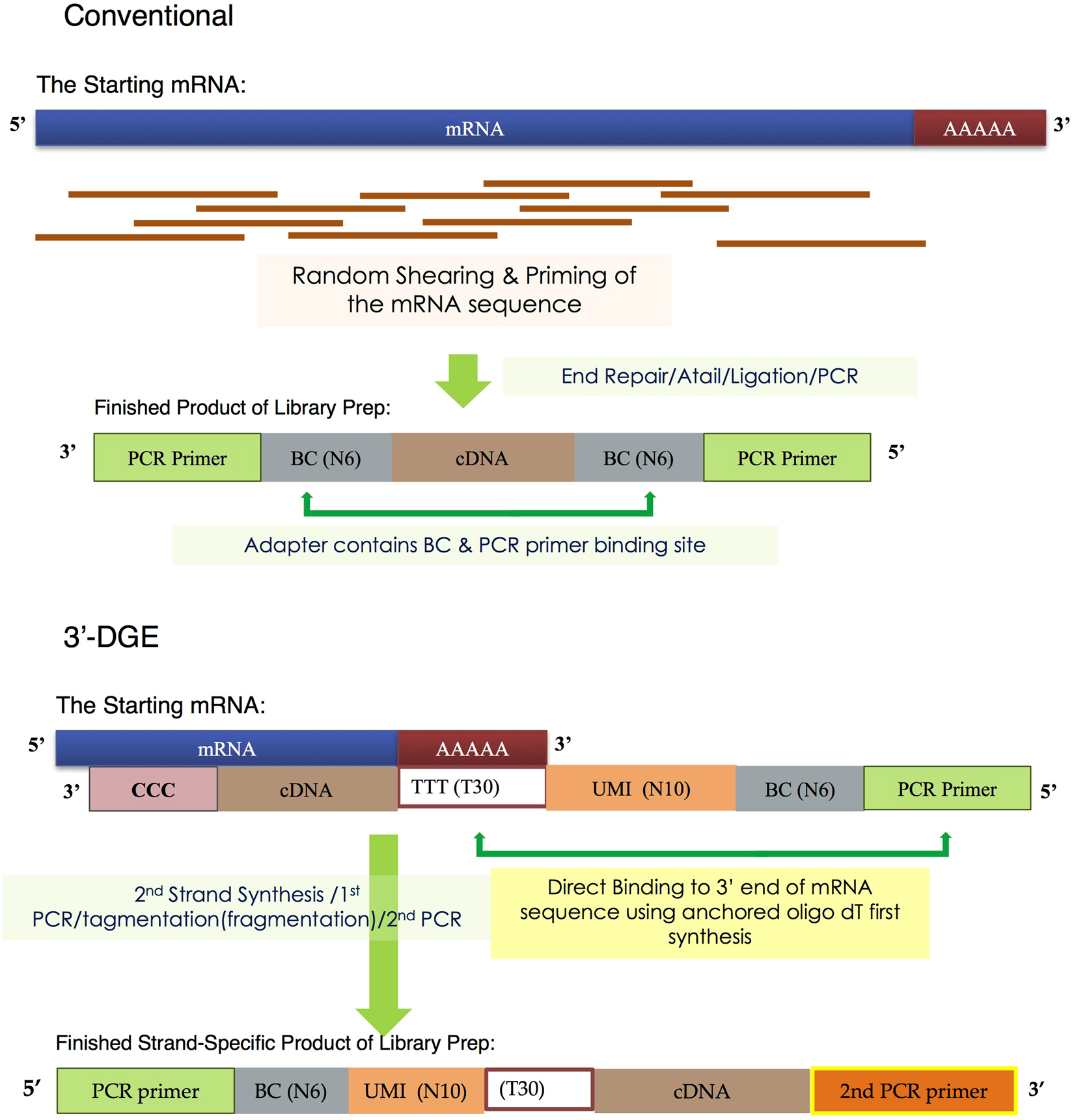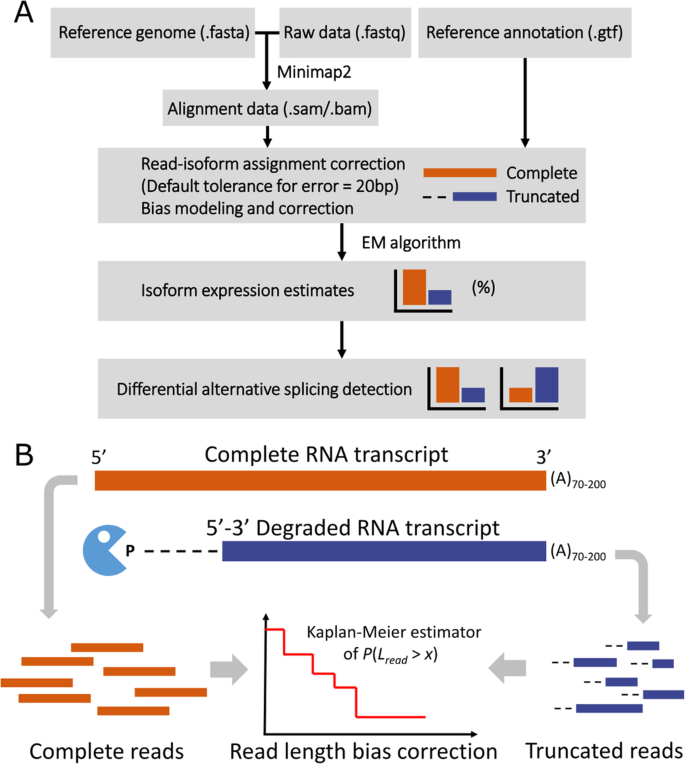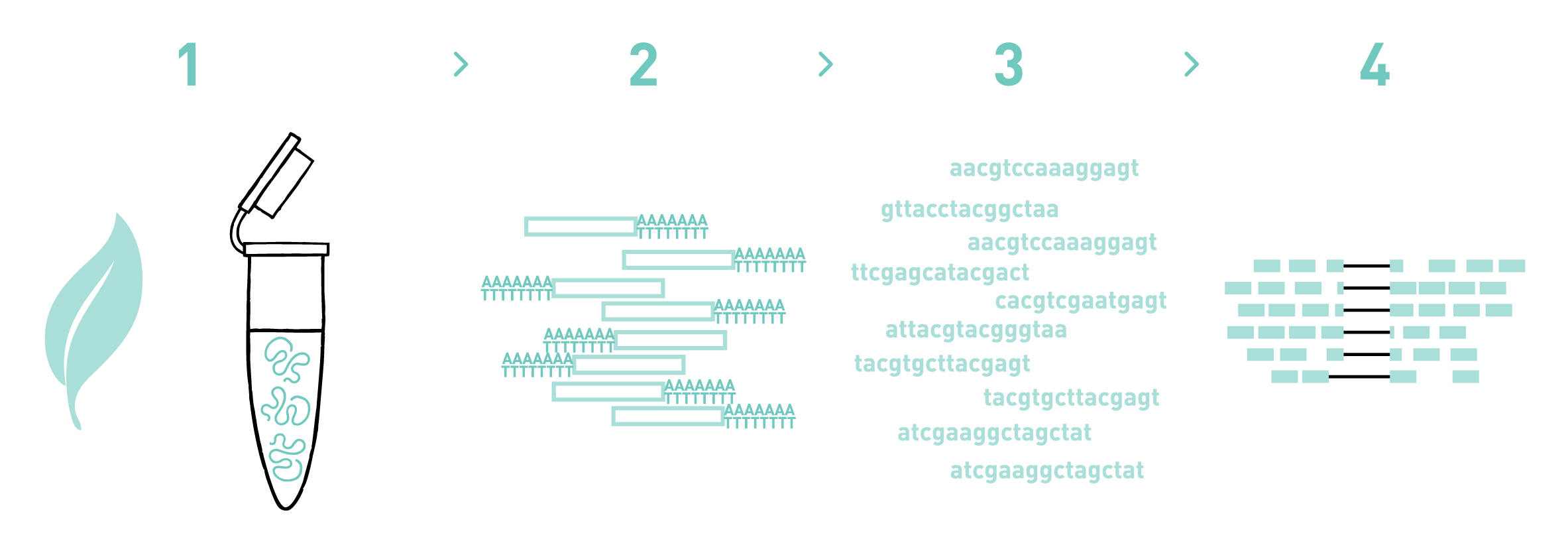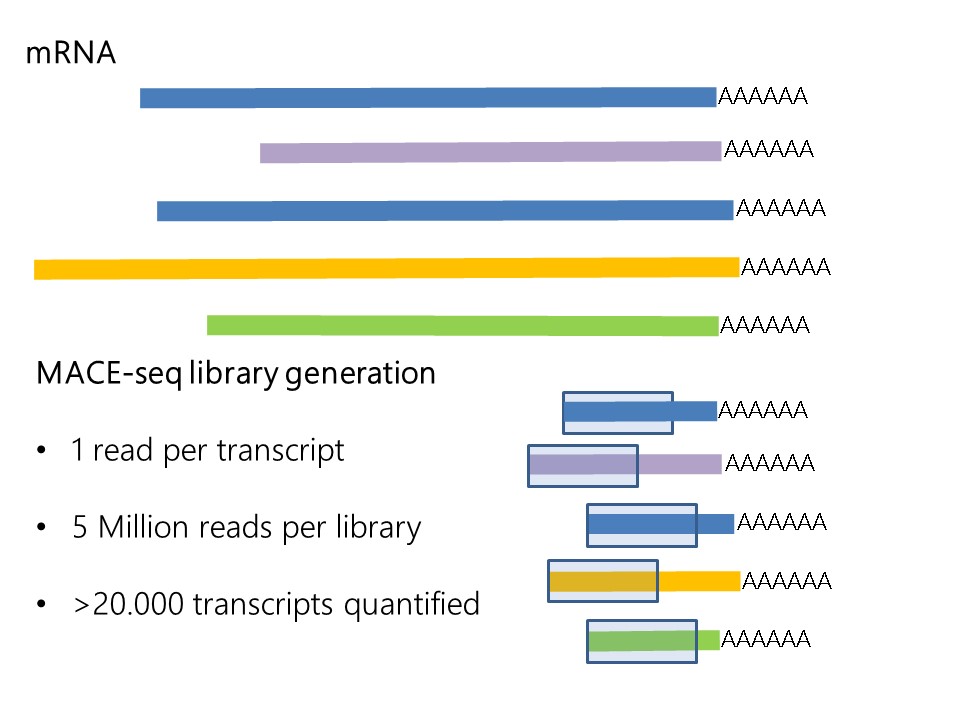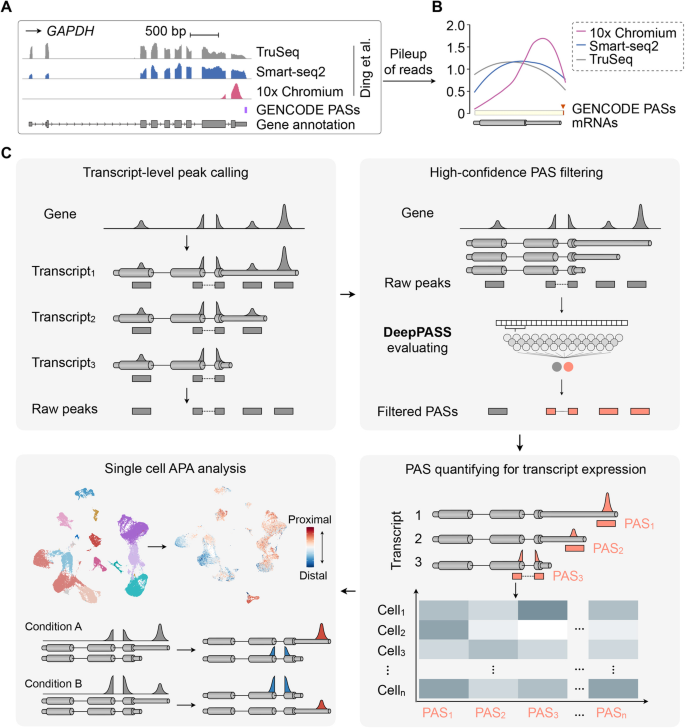
SCAPTURE: a deep learning-embedded pipeline that captures polyadenylation information from 3′ tag-based RNA-seq of single cells | Genome Biology | Full Text

UMI incorporation into small RNA-seq. a Overall workflow. The method... | Download Scientific Diagram

A work flow for 3'end-seq and PacBio Iso-seq. (A) Multiple versions of... | Download Scientific Diagram
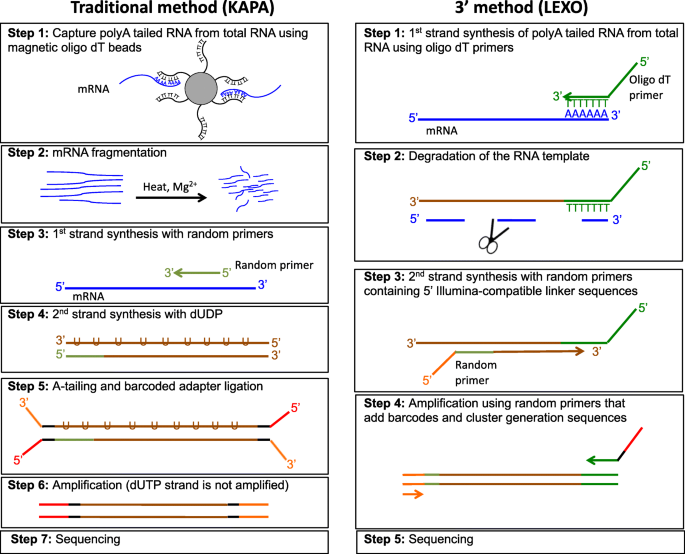
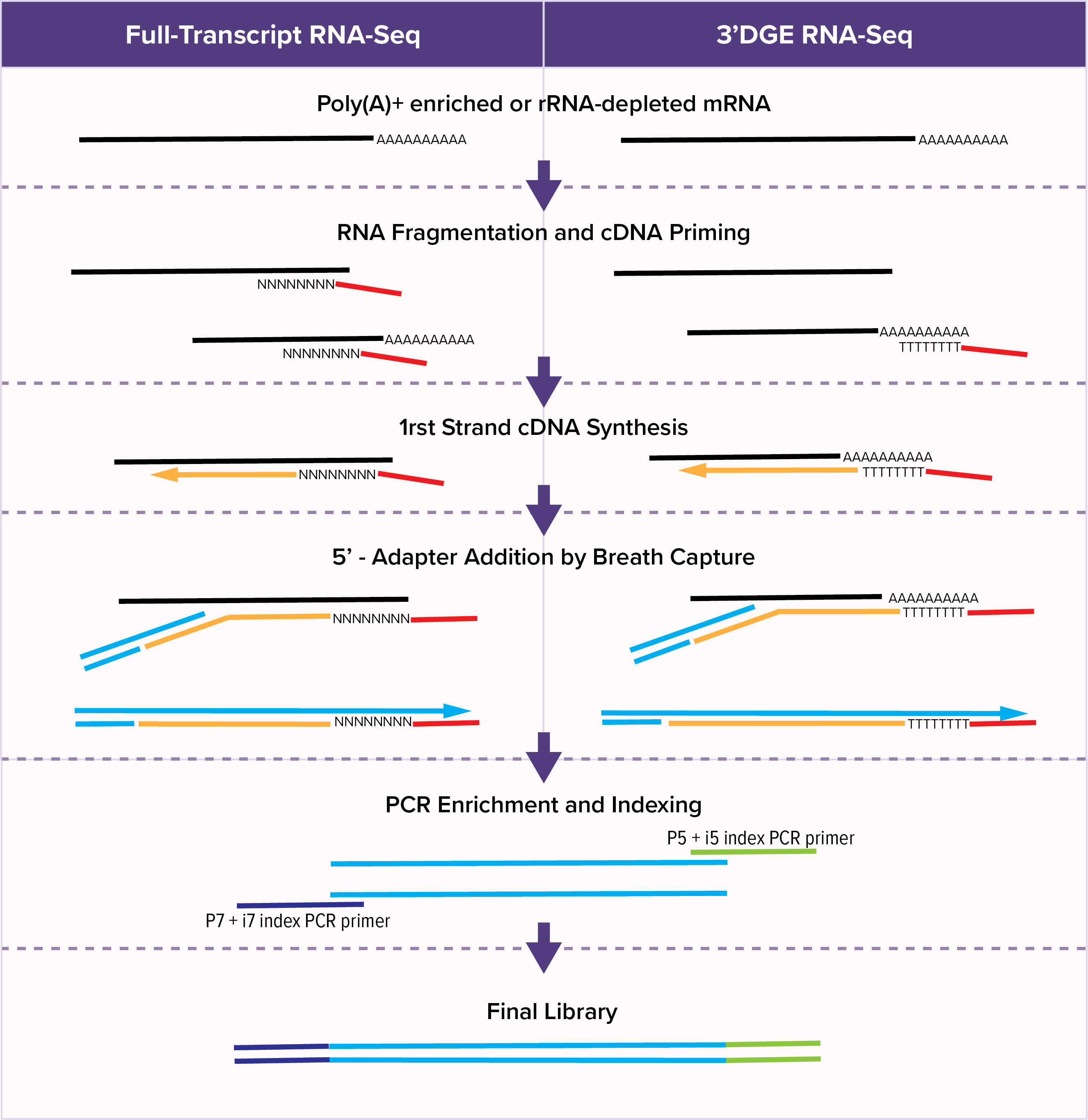
A comparison between whole transcript and 3' RNA sequencing methods using Kapa and Lexogen library preparation methods | BMC Genomics | Full Text
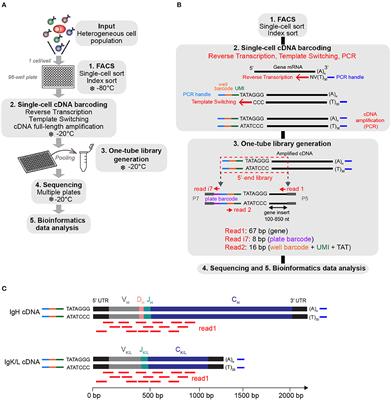
Frontiers | FB5P-seq: FACS-Based 5-Prime End Single-Cell RNA-seq for Integrative Analysis of Transcriptome and Antigen Receptor Repertoire in B and T Cells
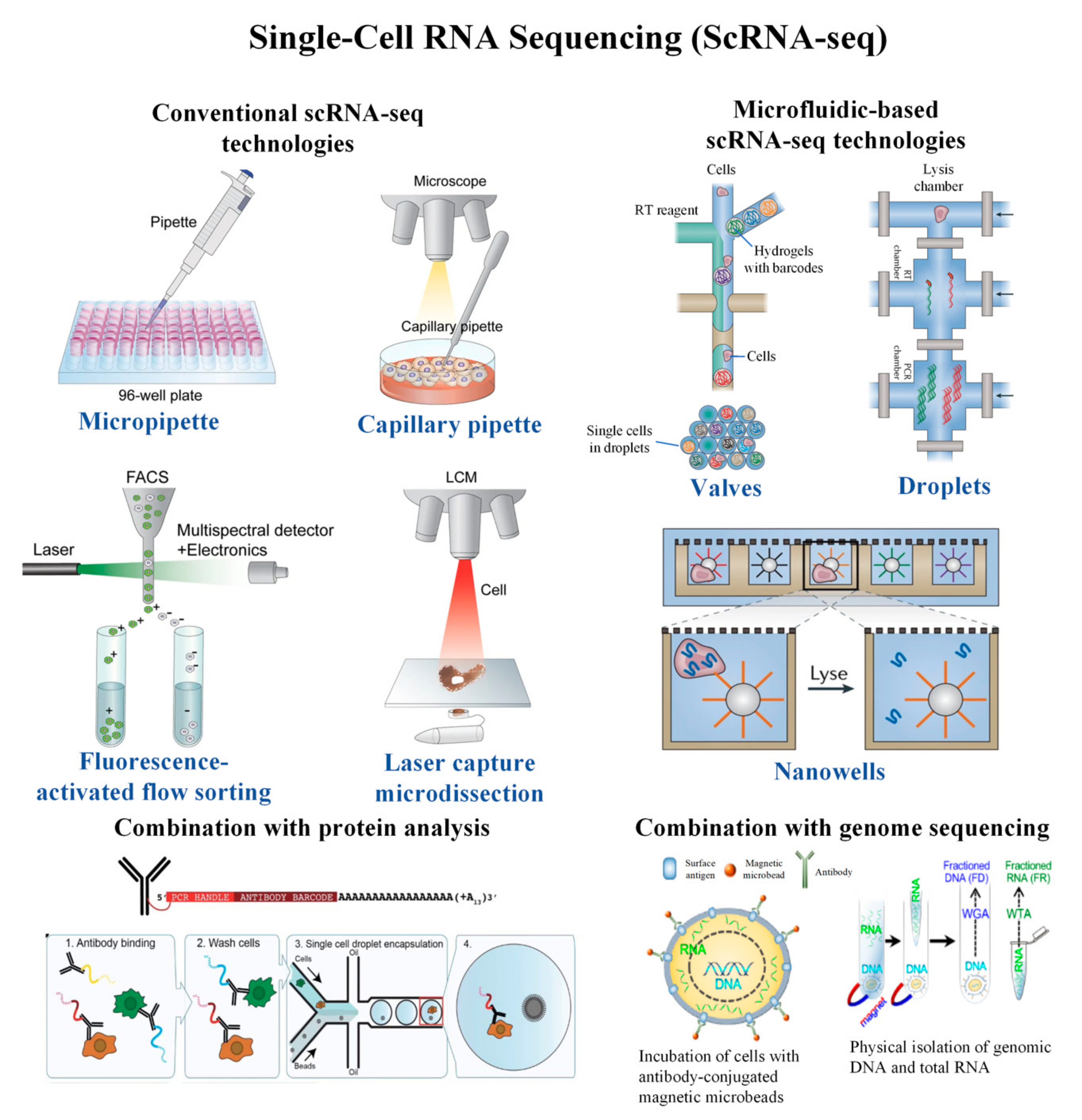
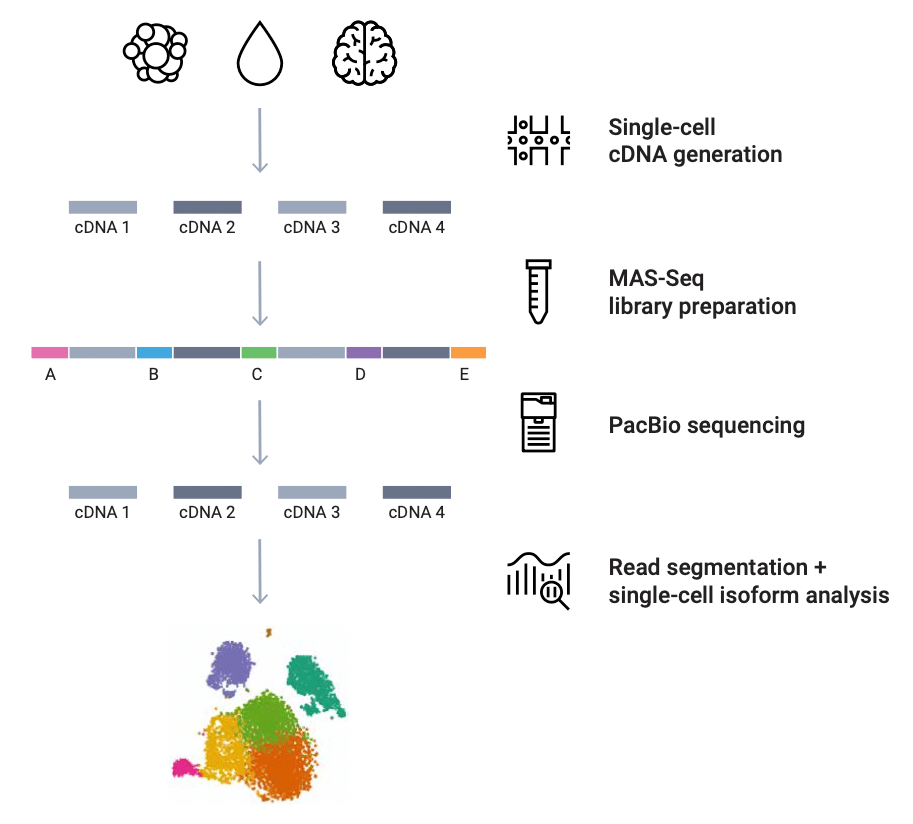
Cells | Free Full-Text | Single-Cell RNA Sequencing and Its Combination with Protein and DNA Analyses